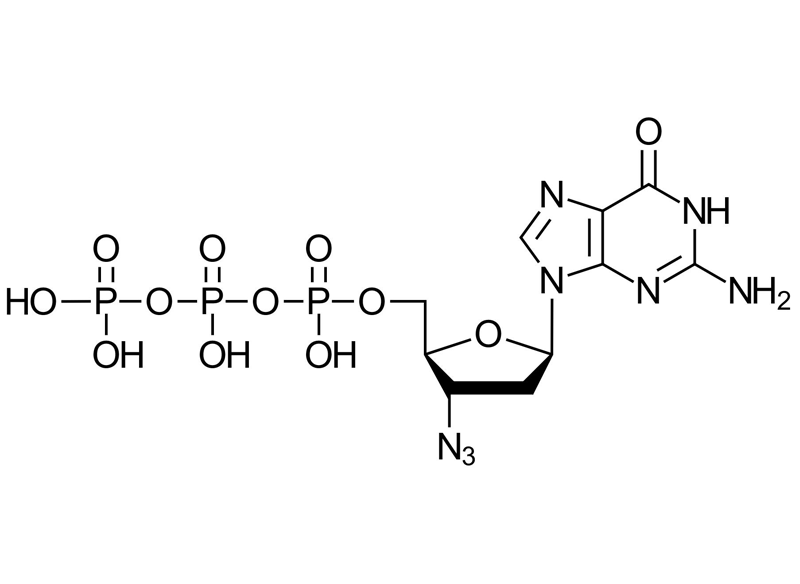
3′-Azido-2′,3′-ddGTP
Triphosphate for modifying of 3'END RNA or ssDNA
-
Azide modified nucleotide closely resemble natural nucleotides and are the basis of chemoenzymatic mRNA labeling. Due to the small size of the azide group, 3′-Azido-2′,3′-ddGTP is well accepted by T7 RNA polymerase and poly(A) polymerase and terminale deoxynucleotidyl transferase (Tdt) for enzymatic incorporation. This is highly beneficial, as labeling of mRNA with our approach is modular and not limited to enzymatic compatibility of bulky pre-labeled nucleotides. Moreover, only a very limited number of such pre-modified nucleotides, mainly dye-labeled ones, are commercially available. Internal and external studies successfully showed e.g. that the poly(A) polymerase adds specifically and sequence independently single azido terminator to the 3’-end of RNA.
The main advantage: Any mRNA can be site-specifically labeled without special requirements or altered production protocols at the 3’-end.
LITERATURE
Site-specific terminal and internal labeling of RNA by poly(A) polymerase tailing and copper-catalyzed or copper-free strain-promoted click chemistry, M. L. Winz et al., 2012, Nucleic Acids Res., Vol. 40, p. 1–13.
https://doi.org/10.1093/nar/gks062
Chemoenzymatic Preparation of Functional Click-Labeled Messenger RNA, S. Croce et al., 2020, ChemBioChem, Vol. 21, p. 1641-1646.
https://doi.org/10.1002/cbic.201900718
Application and design considerations for 3′-end sequencing using click-chemistry, M. K. Jensen et al., 2021, Methods in Enzymology, Vol. 655, p. 1-23.
https://doi.org/10.1016/bs.mie.2021.03.012
Nanopore ReCappable sequencing maps SARS-CoV-2 5′ capping sites and provides new insights into the structure of sgRNAs, C. Ugolini et al., 2022, Nucleic Acids Research, Vol 50(6), p. 3475–3489.
https://doi.org/10.1093/nar/gkac144
Identification of high-confidence human poly(A) RNA isoform scaffolds using nanopore sequencing, L. Mulroney et al., 2022, RNA, Vol. 28(2), p. 162-176.
-
-
Molecular Formula
C10H15N8O12P3
-
Shelf Life
12 months unopened after receipt
-
Storage Conditions
-20 °C
-
Molecular Weight
532.20 g/mol
-
Purity
≥ 95% (HPLC)
-
Physical State
100 mM clear aquaeous solution; colorless
-
CAS Number
94059-38-8 (free acid)
-
Absorption (max)
λmax = 252 nm
-
Ɛ (max)
13,700 cm-1M-1
-
Molecular Formula

