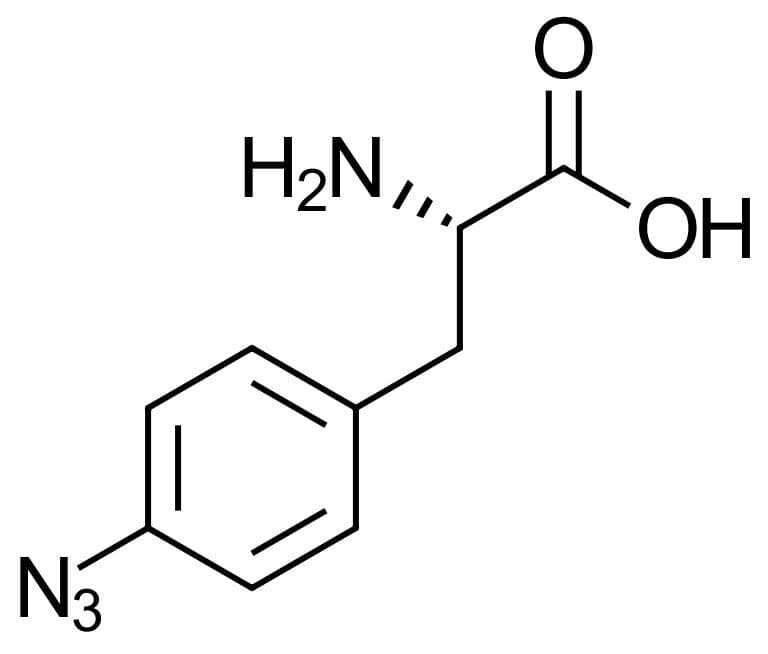
4-Azido-L-phenylalanine
Unnatural azido-modified amino acids for monitoring protein synthesis
-
This unnatural L-phenylalanine derivative, 4-Azido-L-phenylalanine, is used e.g. for genetic code expansion in protein biosynthesis as it can be incorporated into the proteins. Due to the azide-moiety, this molecule allows for specific bioconjugation of various functionalities such as e.g. PEGs, antibodies or fluorescent labels, by click chemistry or Staudinger ligation.
LITERATURE
Cell-free co-production of an orthogonal transfer RNA activates efficient site-specific non-natural amino acid incorporation, C. Albayrak et al., 2013, Nucleic Acids Res., Vol. 41(11), p. 5949-5963.
https://doi.org/10.1093/nar/gkt226
An Expanded Eukaryotic Genetic Code, J. W. Chin et al., 2003, Science, Vol. 301(5635), p. 964-967.
https://www.science.org/doi/10.1126/science.1084772
Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome, H. Neumann et al., 2010, Nature, Vol. 464(7287), p. 441-444.
https://doi.org/10.1038/nature08817
The Locational Impact of Site-Specific PEGylation: Streamlined Screening with Cell-Free Protein Expression and Coarse-Grain Simulation, K. M. Wilding et al., 2018, ACS Synthetic Biology, Vol. 7(2), p. 510-521.
https://doi.org/10.1021/acssynbio.7b00316
Recoded organisms engineered to depend on synthetic amino acids, A. J. Rovner et al., 2015, Nature, Vol. 518(7537), p. 89-93.
https://doi.org/10.1038/nature14095
Uncovering the Domain-Specific Interactome of the TAF1 Tandem Reader Using Site-Specific Azide-Acetyllysine Photochemistry, Y. Yadav et al., 2023, Biochemistry, Vol. 62(2), p. 270–280.
https://doi.org/10.1021/acs.biochem.2c00140
Characterization and Scaled-Up Production of Azido-Functionalized Silk Fiber Produced by Transgenic Silkworms with an Expanded Genetic Code, H. Teramoto et al., 2019, Int. J. Mol. Sci., Vol. 20(3), p. 616.
https://doi.org/10.3390/ijms20030616
Genetic encoding of non-natural amino acids in Drosophila melanogaster Schneider 2 cells, T. Mukai et al., 2010, Protein Science, Vol. 19(3), p. 440-448.
FAQ
-
Is it possible to generate azide- or alkyne-modified peptides?
With modified amino acids azide- or alkyne-modified peptides can be prepared by solid-phase synthesis.
-
How can de novo protein biosynthesis be monitored?
De novo protein biosynthesis can be monitored by feeding of metabolite analogues (so-called metabolic labeling) and subsequent click reaction. Azido-homoalanine for example is recognized as a methionine analogue and is incorporated into de novo synthesized proteins in methionine-free medium conditions. The resulting proteins contain azide moieties and thus can be detected after click to an alkyne-containing reporter molecule (e.g. a fluorescent dye). This non-radioactive method has major practical advantages compared to traditional 35S amino acid incorporation methods.
Alternatively, O-propargyl-puromycin is efficiently incorporated into proteins during de novo protein biosynthesis and can be used in complete medium. The resulting alkyne protein fragments can be detected via click to azide-containing reporter molecules. -
What click conditions should be used for protein click reactions?
A catalyst system based on CuSO4 and sodium ascorbate is recommended in combination with dye azides to label alkyne-modified proteins. Please also refer to our general Click protocols for more details.
Due to the 20 (21) amino acids that are the building blocks of proteins, the physicochemical properties of proteins are more diverse compared to oligonucleotides, which are just composed of 5 major building blocks. Therefore, finding the optimal click conditions is more difficult compared to oligonucleotides and labeling rates are usually lower. Please note that despite these difficulties detection applications (e.g. de novo protein biosynthesis detection) are easily feasible.
-
Is it possible to generate azide- or alkyne-modified peptides?
-
-
Molecular Formula
C9H10N4O2
-
Shelf Life
12 months unopened after receipt
-
Storage Conditions
-20 °C, dry
-
Molecular Weight
206.2 g/mol
-
Purity
≥ 98% (HPLC)
-
Physical State
off-white to brownish powder
-
CAS Number
33173-53-4
-
Additional name
4-N3-L-phenylalanine; H-L-Phe(4-N3)-OH; H-L-4-azido-Phe-OH; H-Phe(4-N3)*HCl, H-4-Azido-Phe*HCl, Phe(pN3)*HCl, para-azido-Phe*HCl, 3-(p-azidophenyl)-L-alanine hydrochloride, 4-azidophenylalanine hydrochloride, p-azido-L-phenylalanine hydrochloride, p-azidophenylalanine hydrochloride
-
Solubility
c = 5% in AcOH 80% (clear slightly colored solution)
-
Molecular Formula

