
DNA-ClickSeq Library Prep Kit
The only NGS prep kit using click chemistry for fragmentation-free, ligation-free DNA libraries

| Size | Catalog No. | Price |
|---|---|---|
| 12 rxn | BCK-DNASeq | € 396,00 |
Chemical Properties
-
Shelf Life
12 months unopened after receipt
-
Storage Conditions
– 20 °C
-
Physical State
kit system made of different components
-
CAS Number
n.a.
-
Preparation/Handling
please see user manual of the kit
Product Information
An improved NGS Workflow with Click Chemistry
Technical overview:
The ClickSeq DNA Library Preparation Kit utilizes the unique chain-terminating properties of 3′-azido-2′,3′-dideoxynucleotides to simplify the construction of DNA sequencing libraries.
As the elongation reaction proceeds, the azido-modified nucleotides are randomly integrated into the growing DNA chain, causing chain termination at the 3′ end. This process generates DNA fragments capped with 3′-azido groups, preventing further elongation.
In the next step, these azido-terminated fragments undergo click-ligation, in which they are covalently linked to alkyne-functionalized sequencing adapters via copper-catalyzed azide–alkyne cycloaddition (CuAAC), also known as click chemistry. This chemical ligation method is both precise and highly efficient, eliminating the need for enzyme-based adapter ligation.
Finally, the adapter-ligated DNA fragments are PCR-amplified to produce high-quality, sequencing-ready libraries. By avoiding artificial fragmentation and enzymatic ligation, ClickSeq offers a cleaner, faster, and more accurate approach to next-generation sequencing (NGS) library preparation.
Standard DNA library preparation typically involves enzyme-based ligation and DNA fragmentation, which can:
- Introducing workflow complexity
- Reduce sample yield
- Cause fragmentation-induced bias
- Generate unwanted byproducts that compromise sequencing accuracy
- Extend processing times
Key Features & Advantages
ClickSeq DNA Library Prep eliminates artificial fragmentation and the need for enzymatic ligation. Instead, it uses a click chemistry-based ligation strategy, ensuring:
- No DNA fragmentation: Preserves genomic structure and reduces workflow complexity.
- Low bias: Improves variant detection and sequencing accuracy.
- Flexible input range: Works with 100 ng to 1 µg of genomic DNA.
- Fast protocol: Entire workflow completed in ~6 hours.
- Ready for Illumina NGS platforms: Includes 12 reactions per kit.
Workflow summary
- Azido-Termination: Genomic DNA is converted to 3′-azido-terminated fragments.
- Click-Ligation: DNA fragments are ligated to alkyne-functionalized adapters using click chemistry.
- PCR Amplification: Adapter-ligated DNA is amplified to generate sequencing-ready libraries.
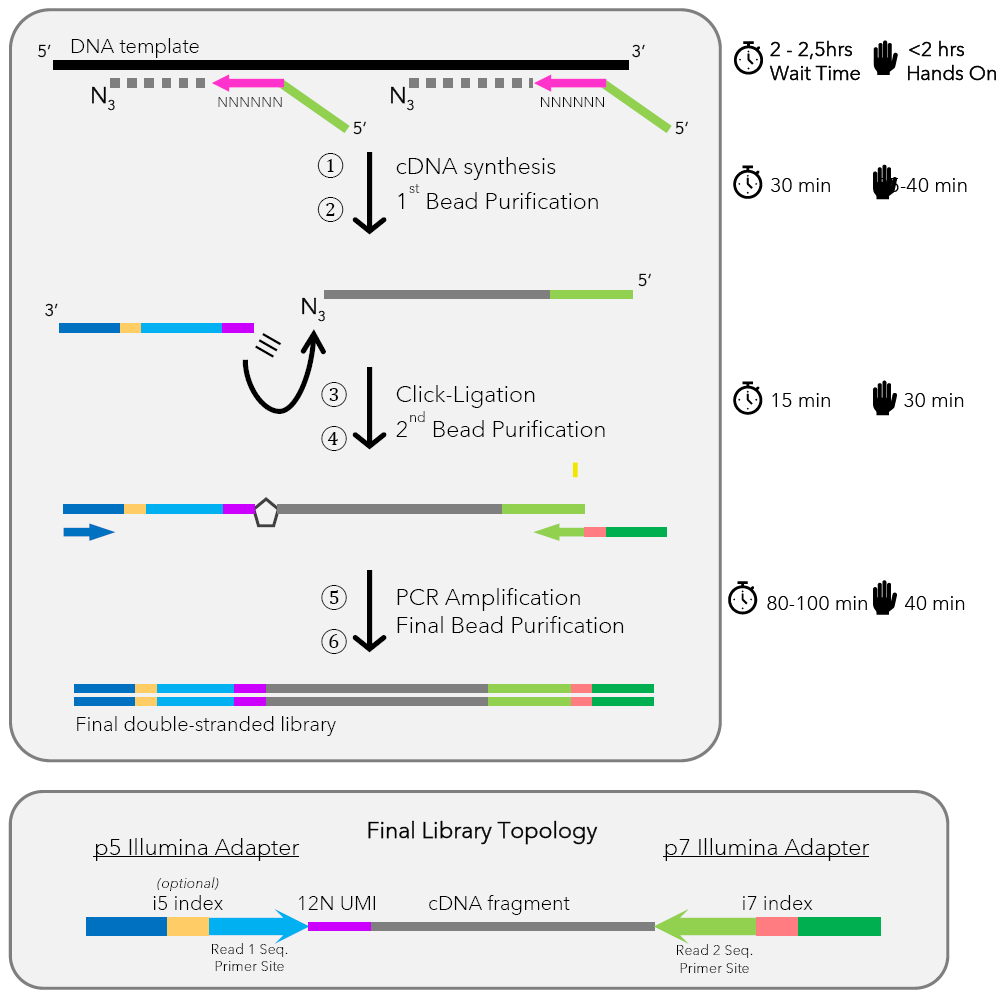
Please note that the DNA-ClickSeq Library Prep Kit does not contain any primers for PCR amplification. A set of 12 compatible i7 index primers is available from baseclick,and is required for DNA-ClickSeq sequencing, unless the user provides their own primers.
Now Compatible for Pangenomic Sequencing
A pangenome is a collection of the common and unique genome parts that are present in a given species. It combines the genetic information of all the genomes sampled, resulting in a large and diverse range of genetic material. This analysis helps understand how genetic variability drives traits within a species and can be applied to various fields like medicine, agriculture, and disease surveillance. This innovative approach is particularly beneficial in several key areas:
- Human Population Genomics: ClickSeq DNA excels in studying genetic diversity and structural variations within human populations. This can provide valuable insights into genetic predispositions, evolutionary biology, and population-specific traits.
- Plant Breeding & Trait Discovery: In the field of agriculture, ClickSeq DNA is instrumental in identifying genetic traits and variations in plants. This aids in breeding programs by enabling the discovery of desirable traits, improving crop yields, and enhancing resistance to diseases and environmental stresses.
- Microbial Comparative Genomics: ClickSeq DNA is also highly effective in comparing genomic differences among microbial species. This enhances our understanding of microbial diversity, pathogenicity, and antibiotic resistance, which is crucial for developing new treatments and understanding microbial evolution.
This innovative approach makes ClickSeq DNA an excellent choice for researchers aiming to explore complex genomic landscapes and gain detailed insights into genetic diversity.
FAQ
-
What is ClickSeq?
ClickSeq is a platform and method for making Next-Generation Sequencing libraries. It is called ‘ClickSeq’ as it replaces the ligations and fragmentation steps common to NGS library preps with ‘Click-Chemistry’.
ClickSeq follows the same principle of all Next Generation Sequencing (NGS) Library Prep Kits that seek to generate short fragments of cDNAs that are flanked with the appropriate sequencing adaptor for the user’s choice of sequencing platform (e.g. Illumina).
The process works by supplementing the Reverse Transcription step of an Next Generation Sequencing (NGS) Library Prep Kit with small amounts of terminating ‘azido-nucleotides’. These are stochastically incorporated into the cDNA during first stranded synthesis, yielding 3’azido-terminated cDNA molecules. The azido group is then ‘Click-Ligated’ to an alkyne-functionalized sequencing adaptor using Click-Chemistry (Copper Catalysed Azide-Alkyne Cycloaddition, CuAAC). The resultant click-linked single-stranded cDNA is then PCR amplified to generate a final sequencing-ready NGS library.
-
How does ClickSeq differ from traditional NGS library prep kits?
Unlike conventional kits that rely on artificial DNA fragmentation and enzymatic ligation, ClickSeq avoids these steps altogether. Instead, it uses click chemistry for adapter ligation, which preserves the entire DNA structure. This approach not only reduces bias but also simplifies the workflow, making the process more efficient and reliable.
-
What input amount of DNA is required for the ClickSeq DNA kit?
The kit is compatible with 100 ng to 1 µg of genomic DNA, making it suitable for a wide range of sample types, including low-input or precious material.
-
Is ClickSeq compatible with all NGS platforms?
The kit is optimized for Illumina sequencing platforms. For other platforms, adapter compatibility would need to be evaluated or customized.
-
Can this kit be used for structural variant analysis or pangenomic studies?
Yes. The absence of fragmentation makes ClickSeq ideal for pangenomic and structural variation analysis, where preserving native DNA architecture is critical.
-
Where can I find a protocol for ClickSeq?
Our protocol can be found here:
https://www.protocols.io/view/clickseq-random-primed-protocol-with-single-indexi-n92ld8jkov5b/v1
-
What do I need in addition to the kit to make a ClickSeq library?
Required third-party reagents include:
- SuperScript III™ Reverse Transcriptase, 200U/µL (Invitrogen; 18080-093 or 18080-044)
- ClickSeq Library Prep Kit Index Primers (baseclick, BCK-RNAseq-IP)
- [optional] RNaseOUT™ Recombinant Ribonuclease Inhibitor, 40U/µL (Invitrogen; 10777)
- OneTaq® 2X Master Mix with Standard Buffer (NEB; M0482S or M0482L) (Note: you must use OneTaq for this step, this enzyme cannot be substituted for a different PCR enzyme)
- [optional] RNase H, 5000 units/mL (NEB; M0297S or M0297L)
- SPRIselect (Beckman Coulter; B23317) or equivalent DNA/RNA Purification Beads (also known as SPRI beads)
- Nuclease free water
- 80% ethanol (made fresh)
-
Where can I find more information?
Please see an extensive set of FAQs:
-
What is included in the ClickSeq DNA Library Prep Kit?
Each kit includes reagents for 12 complete library prep reactions, including azido-nucleotide mix, alkyne-functionalized adapters, and buffers optimized for click-ligation and PCR.

