EdU T Cell Proliferation Assay for Flow Cytometry
ClickTech EdU T Cell Proliferation Kit for Flow Cytometry

| Size | Catalog No. | Price |
|---|---|---|
| Dye 488 / 48 Assays | BCK-TCell-FC488_48 | € 150,00 |
| Dye 488 / 192 Assays | BCK-TCell-FC488_192 | € 400,00 |
| Dye 555 / 48 Assays | BCK-TCell-FC555_48 | € 150,00 |
| Dye 555 / 192 Assays | BCK-TCell-FC555_192 | € 400,00 |
| Dye 647 / 48 Assays | BCK-TCell-FC647_48 | € 150,00 |
| Dye 647 / 192 Assays | BCK-TCell-FC647_192 | € 400,00 |
-
Cell analysis – T Cell Proliferation Assays for Flow Cytometry
Cell analysis allows scientists to gain unique insights into cell proliferation, viability, and toxicity, enabling the monitoring of cell response and cell health in cultures after treatment with various stimuli.
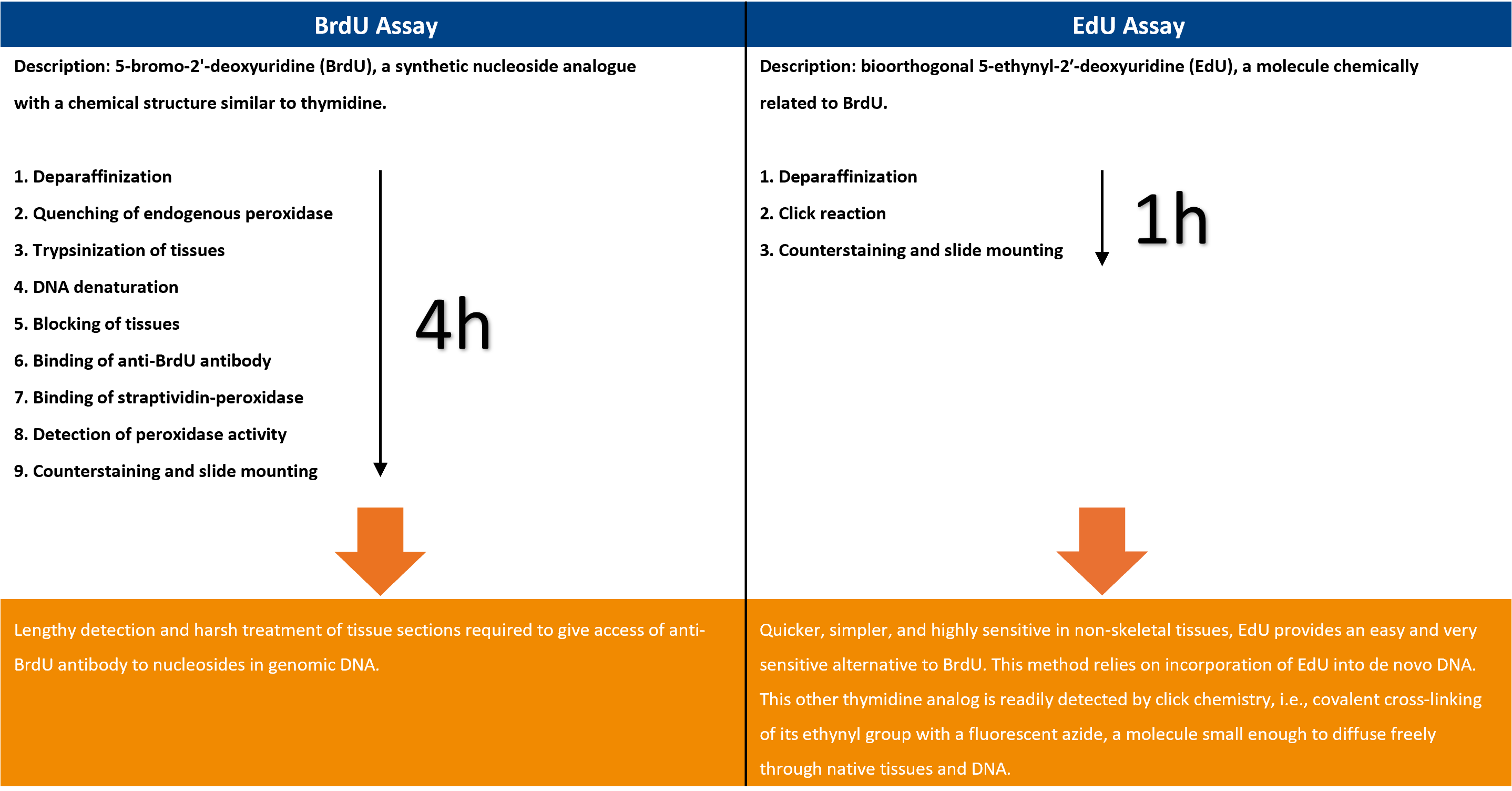
Detection of DNA replication is a powerful tool for studying proliferation of both cell populations and individual cells. DNA replication can be detected through the incorporation of a nucleoside analog such as BrdU or EdU into DNA. The detection of these two thymidine analogs differs significantly and has implications for test results, potential applications, and multiplexing capability. Compared to other cell proliferation methods, the EdU proliferation assay does not involve radioactive isotopes or antibodies to detect newly synthesized DNA.
The ClickTech EdU cell proliferation assay enables estimates of cell proliferation rates by determining the fraction of cells replicating their DNA.
Schematic representation of EdU cell proliferation assays. A) Incubation of cells with EdU. B) EdU is incorporated during active DNA synthesis. C) Detection of cell proliferation via click chemistry, wherein the use of a range of different fluorescent dyes is possible.
ClickTech EdU T Cell Proliferation Assays for FC
ClickTech EdU T cell Proliferation assay enables monitoring genotoxicity, anticancer drug evaluation or cell cycle analysis by direct detection of DNA replication in proliferating cells. The technology is based on the labeling of DNA with EdU (5-ethynyl-2′-deoxyuridine), an alkyne-modified nucleoside that is incorporated into the replicating DNA of living cells and its conjugation to a fluorochrome in situ. This nucleoside contains a reactive alkyne group (instead of the bromine atom in BrdU) and can be conjugated to molecules bearing azide groups through a click reaction. After an incubation period that depends on the specific cell type, the cells are washed, fixed and permeabilized to allow the detection reagents to penetrate.
The detection is then carried out via the copper-catalyzed click reaction (also known as Azide-Akyne Cyclo-addition or CuAAC). Click reactions show high quantitative yields, run under mild conditions and are easy to perform. In order to start the reaction, a click cocktail, containing buffer, catalytic reagents and a fluorescent dye, is added to the cells. The dye contains an azide group that reacts with the alkyne group of the incorporated EdU. Only cells that actively replicate their DNA during the EdU pulse can be detected by this method.
Functional alkyne and azide groups do not occur in natural cell systems and do not react without catalytic reagents. This ensures that the EdU cell proliferation method, based on click chemistry, is highly selective, sensitive and reproducible. This technology allows an almost unlimited choice of different and easy to synthesize fluorescent dyes and thus offers maximum flexibility in terms of “detection colors” and analytical devices. baseclick currently offers EdU T Cell proliferation assays for flow cytometry and three standard dyes (488, 555, 647 nm).
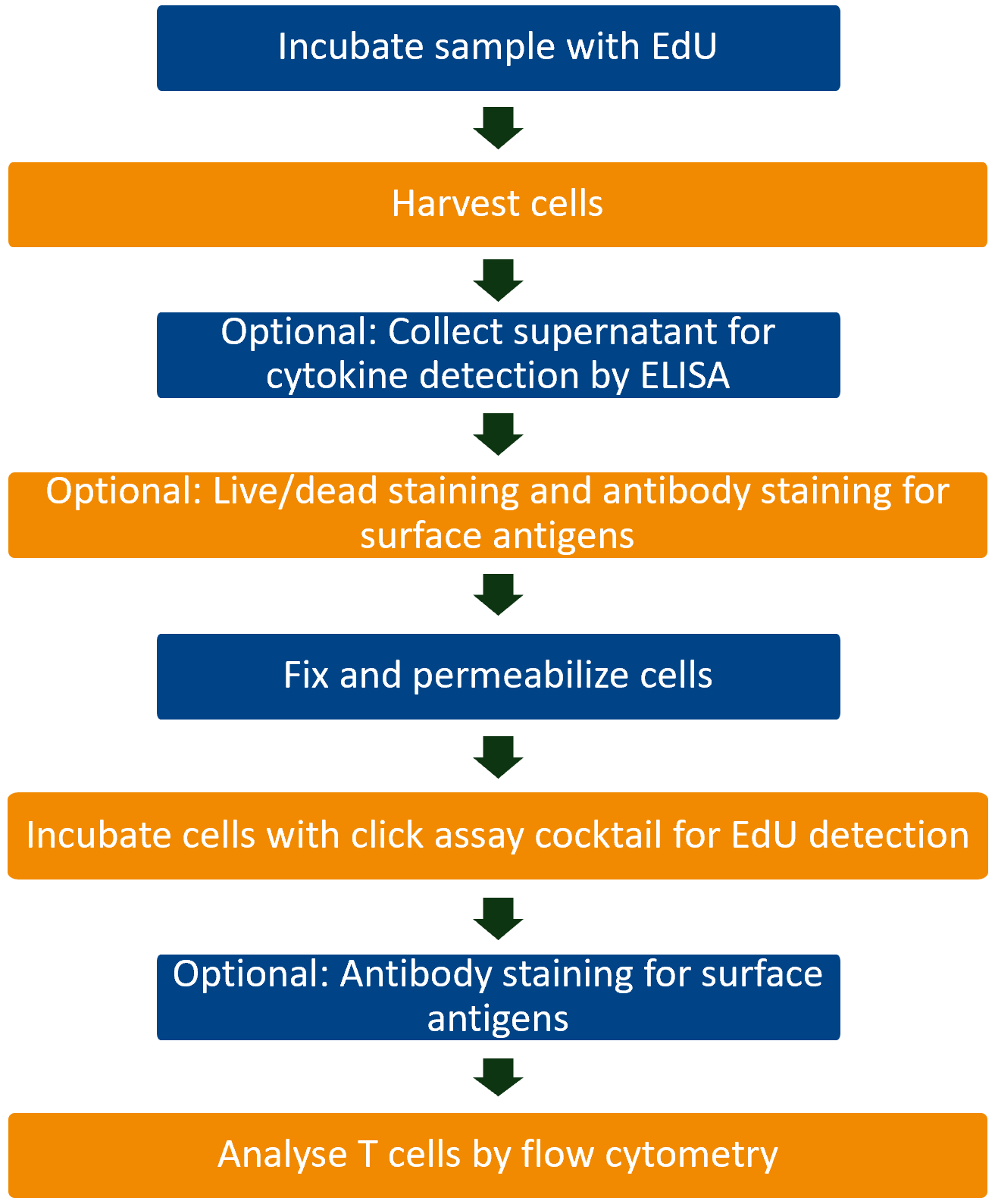
Workflow
Advantages:
- Increased selectivity and sensibility in comparison to BrdU
- Simple, fast and reliable assay
- High sensitivity paired with very low toxicity
- A cost-effective assay
- Multiplexing possibility
Application
Comparison between the BrdU and the EdU assay:
Comparison between cell tracing and the EdU assay:
Fluorescent cell tracing reagents exhibits several limitations as it can reduce T cell proliferation, viability, responsiveness, and cell proliferation can only be determined after completion of full rounds of cell division.
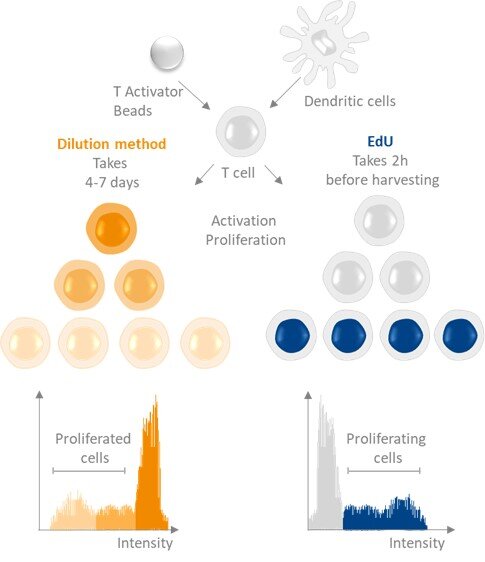
Stimulation of human T cell activation via comparing the dilution-based T cell proliferation probe CellTrace Far Red and EdU. ClickTech EdU T cell proliferation Kit overcomes the limitations of dilution-based probes, thus providing a superior alternative for measuring T cell proliferation.
Although both the ClickTech EdU Cell Proliferation assays and fluorescent cell tracing assays such as CellTraceTM are used to estimate cell proliferation, the actual information they provide is different as they measure distinct parameters/aspects of cell proliferation:
- By measuring dilution of a signal through cell division, fluorescent cell tracing assays give an estimate of how many cell generations have taken place between the labelling pulse with the tracing reagent and the time at which signal detection by FACS is performed (usually 4-5 days).
- In contrast, by determining the fraction of cells replicating their DNA, the ClickTech EdU T cell Proliferation assay estimates the proliferation rate of a T cell population at the time of the EdU pulse, which in the standard application of the assay is immediately before detection. Changes in the proliferation rate of a T cell population over a prolonged period can also be monitored by performing the ClickTech EdU T cell Proliferation assay at different time points during that period. Information on the number of cell generations that take place over a period can also be obtained by applying a single, initial EdU pulse on a cell population and performing EdU detection on aliquots of the same cell population not only immediately after the EdU pulse, but also at later time points after washout of the pulse (a so-called chase). Here too, the decrease of signal intensity allows estimating how many cell divisions have occurred since the initial pulse. Thus, the ClickTech EdU T cell Proliferation assay is more versatile than fluorescent cell tracing assays.
LITERATURE
A sensitive and less cytotoxic assay for identification of proliferating T cells based on bioorthogonally-functionalized uridine analogue, F. C. Stempels et al., 2022, Journal of Immunological Methods, Vol. 502, p. 113228.
https://doi.org/10.1016/j.jim.2022.113228FAQ
-
How does EdU labeling compare to methods based on label dilution?
Mostly, dilution-based T cell proliferation probes conjugate unspecifically to amine groups in cells. Each cell division then results in dilution (takes several days) of the probes. Main disadvantages are cytotoxicity, an impact on T cell activation and long incubation time (several days and up to a week) before readout. The superior EdU cell proliferation assay relies on the incorporation of the modified uridine analog EdU in the DNA of proliferating T cells (just 2 hours before harvesting) and subsequent detection. Both methods enable detection and quantification of T cell proliferation by flow cytometry. Our EdU incorporation assay is very sensitive, and can be multiplexed with other fluorescent stainings.
- EdU outperforms a standard dilution-based probe in detecting T cell proliferation
- The EdU assay is less cytotoxic for human T cells
- The EdU assay offers superior signal-to-background ratio
- The EdU assay allows for better discernable interferon gamma responses
-
Can I combine the EdU T cell Proliferation assay and staining with PerCP, APC, APC-based tandems, RPE, PR-tandem, Quantum Dot antibody conjugates or intracellular antigens?
Yes, this is feasible. Please note that staining with PerCP, APC and APC-based tandems may be performed before the EdU detection step (click reaction), while RPE, PR-tandem or Quantum Dot antibody conjugates and detection of intracellular antigens should be performed after the click reaction. Check also the user manual for further information. Please, make sure that the emission spectra of the fluorochromes to be combined do not overlap extensively, so that their signals can be distinguished unequivocally using appropriate emission filters or spectral demixing.
-
When may break points be introduced during the protocol?
It is possible to safely interrupt the protocol after the fixation step. Thereto, remove the fixation solution and wash as suggested by the user manual, then the cells can be stored in buffer at 4° C. Alternatively, the experiment can also be safely interrupted after permeabilization, again as described above.
Please note: It is important to proceed with the experiment if the click cocktail for the detection of the EdU has been prepared already. -
How to determine the EdU incubation time?
The EdU incubation time depends on the cell type or organism, the applied EdU concentration and the experimental design. For a start it is advisable to refer to a literature protocol (which is close to your experimental setup) and to test the conditions with a low number of samples. As a general guideline we recommend to use a maximum of 10 µM final EdU in the cell culture medium for incubations. For longer incubation (> 1 day) the concentration should be decreased to 1-5 µM.
-
-
Shelf Life
12 months unopened after receipt
-
Storage Conditions
2-8 °C
-
Physical State
kit system made of different components
-
CAS Number
n.a.
-
Excitation (max)
Dye 488: 496 nm | Dye 555: 546 nm | Dye 647: 643 nm
-
Emission (max)
Dye 488: 516 nm | Dye 555: 579 nm | Dye 647: 662 nm
-
Ɛ (max)
Dye 488: 83.000 cm-1M-1 | Dye 555: 91.000 cm-1M-1 | Dye 647: 250.000 cm-1M-1
-
Preparation/Handling
please see user manual of the kit
-
Shelf Life

