
EdU Cell Proliferation Assay for Flow Cytometry
ClickTech EdU Cell Proliferation Kit for Flow Cytometry
| Size | Catalog No. | Price |
|---|---|---|
| Dye 488 / 50 Assays | BCK-EdU488FC50 | € 495,00 |
| Dye 488 / 100 Assays | BCK-EdU488FC100 | € 770,00 |
| Dye 555 / 50 Assays | BCK-EdU555FC50 | € 495,00 |
| Dye 555 / 100 Assays | BCK-EdU555FC100 | € 770,00 |
| Dye 594 / 50 Assays | BCK-EdU594FC50 | € 495,00 |
| Dye 594 / 100 Assays | BCK-EdU594FC100 | € 770,00 |
| Dye 647 / 50 Assays | BCK-EdU647FC50 | € 495,00 |
| Dye 647 / 100 Assays | BCK-EdU647FC100 | € 770,00 |
Chemical Properties
-
Shelf Life
12 months unopened after receipt
-
Storage Conditions
2-8 °C
-
Physical State
kit system made of different components
-
CAS Number
n.a.
-
Excitation (max)
Dye 488: 496 nm | Dye 555: 546 nm | Dye 594: 584 nm | Dye 647: 643 nm
-
Emission (max)
Dye 488: 516 nm | Dye 555: 579 nm | Dye 594: 603 nm | Dye 647: 662 nm
-
Ɛ (max)
Dye 488: 83.000 cm-1M-1 | Dye 555: 91.000 cm-1M-1 | Dye 594: 110.000 cm-1M-1 | Dye 647: 250.000 cm-1M-1
-
Preparation/Handling
please see user manual of the kit
Product Information
Fast, Accurate, and Antibody-Free DNA Synthesis Detection
Cell based analysis: Cell Proliferation Flow Cytometry Assays
The ClickTech EdU Cell Proliferation Kit for Flow Cytometry provides a robust and efficient method for quantifying de novo DNA synthesis during cell proliferation. Based on the incorporation of 5-ethynyl-2′-deoxyuridine (EdU) into replicating DNA, this assay eliminates the need for DNA denaturation and antibody-based detection required by BrdU methods. Detection is achieved via copper-catalyzed azide-alkyne cycloaddition (click chemistry) a highly specific and mild reaction recognized with the 2022 Nobel Prize in Chemistry.

Comparison of Cell Proliferation Assays
The table below summarizes key differences among commonly used cell proliferation assays. EdU-based detection using ClickTech stands out for its high sensitivity and specificity, rapid processing time (~30 minutes), and compatibility with multiplexing, without the need for harsh DNA denaturation or radioactive materials.
| Assay Type | Detection Methods | Sensitivity | Specificity | Processing Time | Toxicity | Multiplexing | Advantages | Limitations |
| EdU (ClickTech) | Click chemistry, fluorescent tags | High | High | ~30 min (post-label) | Low | Yes | Precise S-phase detection, no denaturation | Requires click chemistry reagents |
| BrdU | Antibody-based, DNA denaturation | Moderate | Moderate | ~2-4 hours | Moderate | Limited | Widely established method | Requires denaturation, longer workflow |
| 3H-thymidine | Radioactive incorporation | High | High | ~24 hours | High | No | Highly sensitive DNA synthesis tracking | Radioactive, disposal hazards |
| CFSE | Dye dilution, fluorescence | High | Moderate | ~1-2 hours | Moderate | Limited | Tracks division history | Dye toxicity, no phase specificity |
| MTT | Metabolic activity, colorimetric | Low | Low | ~4 hours | Low | No | Simple, no labeling required | Indirect measure, affected by cell death |
In contrast, BrdU assays, while widely established, require longer workflows and denaturation steps, and ³H-thymidine involves radioactive hazards and extended processing. CFSE offers division tracking but lacks phase specificity and can introduce dye toxicity, while MTT provides only an indirect measure of proliferation based on metabolic activity. This comparison highlights EdU as the most efficient and versatile option for modern flow cytometry applications.

Kit Features
This kit includes complete set of reagents and an optimized protocol for flow cytometry-based proliferation analysis
It supports multiparametric analysis, allowing seamless combination with cell cycle dyes, fluorescent proteins, and immunostaining markers.
- EdU reagent
- Fluorescent azide dye (e.g., 6-FAM, TAMRA, Sulforhodamine and Eterneon red)
- Reaction buffer and optional copper catalyst
- Fixation and permeabilization solutions
- Detailed protocol
Available in 50- and 100-assay formats for flow cytometry applications.
Performance Demonstration
To illustrate the sensitivity and reliability of the ClickTech EdU Cell Proliferation Kit, we conducted a flow cytometry assay using HeLa cells. This example highlights the kit’s ability to clearly distinguish proliferating cells from non-proliferating controls, delivering precise and reproducible fluorescence signals within a streamlined workflow.
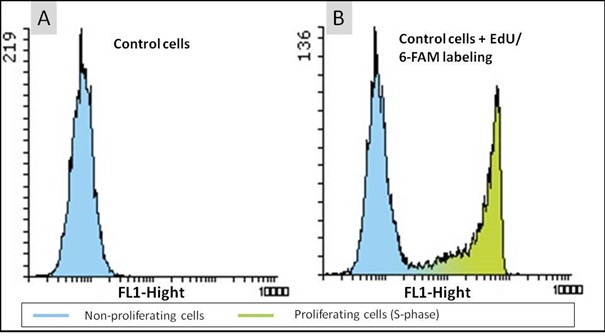
Figure 2: Cell proliferation assay with the EdU Cell Proliferation Kit for Flow Cytometry (BCK-EdU488FC). HeLa cells were either untreated (A) or incubated with 10 µM EdU for 2 hours (B). The click reaction was performed using 6-FAM Azide and fluorescence intensity of 10.000 cells was measured by flow cytometry. The results are presented in form of histograms, showing the cell number in the y-axis and the FL1-Fluorescence in the x-axis.
Advantages of EdU-Based Flow Cytometry
- Direct detection of DNA synthesis without antibodies and DNA denaturation
- Preserves cell structure, enabling downstream immunostaining
- Fast /simple protocol and fluorescence detection in ~60 minutes
- Allows multiplexing with cell cycle dyes, fluorescent proteins such as GFP, RFP, phycobiliproteins
- Compatible with multiple fluorophores (488, 555, 594, 647 nm) for multiparametric analysis
The EdU-based assay is non-toxic to cells during incorporation and offers a safer alternative to dye-dilution methods such as CFSE.
Applications
- Monitors genotoxicity and assesses the impact of environmental or chemical stressors on cell division.
- Evaluates anticancer drug efficacy by measuring proliferation inhibition.
- Analyzes cell cycle dynamics across diverse cell types, including T-cells, HeLa, and primary cells.
- Supports immune response studies, such as T-cell proliferation post-activation.
- Enables high-throughput screening for drug discovery and cytotoxicity assessment.
LITERATURE
- Cytolethal Distending Toxin Promotes Replicative Stress Leading to Genetic Instability Transmitted to Daughter Cells, W. Tremblay et al., 2021, Frontiers in Cell and Developmental Biology, Vol. 9, p. 656795. https://doi.org/10.3389/fcell.2021.656795
- Innovative DNA-Targeted Metallo-prodrug Strategy Combining Histone Deacetylase Inhibition with Oxidative Stress, T. McGivern et al., 2018, Mol. Pharmaceutics, Vol.15, p. 5058-5071. https://doi.org/10.1021/acs.molpharmaceut.8b00652
- YB-1 Expression and Phosphorylation Regulate Tumorigenicity and Invasiveness in Melanoma by Influencing EMT, C. Kosnopfel et al., 2018, Mol Cancer Res., Vol. 16, p. 1149-1160. https://doi.org/10.1158/1541-7786.MCR-17-0528
- Chronic exposure to Cytolethal Distending Toxin (CDT) promotes a cGAS-dependent type I interferon response, B. J. Pons et al., 2021, Cellular and Molecular Life Sciences, Vol. 78, p. 6319–6335. https://doi.org/10.1007/s00018-021-03902-x
- Protocol for cell proliferation and cell death analysis of primary muscle stem cell culture using flow cytometry, Garcia P., et al., 2024, STAR Protocols 5:4. https://doi.org/10.1016/j.xpro.2024.103411
- Analysis of Cell Proliferation and Homeostasis Using EdU Labeling, Flomerfelt F.A., Gress R.E., 2016, Methods Mol. Biol, 1323:211–220. https://doi.org/10.1007/978-1-4939-2809-5_18
- An EdU-based flow cytometry assay to evaluate chicken T lymphocyte proliferation, Alvarez K.L., et al., 2020, BMC Vet. Res. 2020 Jul 6;16:230. https://doi.org/10.1186/s12917-020-02433-0
FAQ
-
What type of cells can incorporate EdU?
The EdU cell proliferation assay has been applied to many different cell types and organisms from prokaryotic to eukaryotic. Cell lines such as E. coli, HeLa, HEK, MOLM are arguably among the most routine applications, but also animals, like mouse, rat, the nematode C. elegans, crickets (Gryllus bimaculatus), chicken (Gallus domesticus) and zebra fish (Danio rerio) or even plants (e.g. Arabidopsis thaliana) can be applied.
Cells that possess a pyrimidine pathway that can phosphorylate EdU to the corresponding triphosphate, which is then accepted by the host DNA polymerase for incorporation into DNA during replication. -
Can I perform EdU cell proliferation detection on living cells?
EdU is incorporated into living cells, but the detection reaction must be performed on fixed and permeabilized samples.
-
How does EdU labeling compare to BrdU or the 3H-thymidine incorporation assay?
All three methods enable to determine cell proliferation directly by incorporation of a metabolite analogue and subsequent detection. The 3H-thymidine incorporation assay is very sensitive, but the radioactive compound requires specialized equipment and dedicated lab space for handling. EdU and BrdU assays are non-radioactive alternatives with decreased risk for health and environment. Compared to the BrdU incorporation assay the EdU assay is more sensitive, requires less handling time and needs no harsh DNA denaturing conditions for detection. Therefore, the EdU cell proliferation is also compatible with multiplexing.
-
Can I combine DAPI staining and EdU detection?
Yes, this is feasible. Please note that DAPI staining should be done after the click detection step. Alternatively, SYBR Green DNA staining can be used. But, please note that SYBR green should not be used with dyes of 488 nm wavelengths.
-
When can I safely interrupt the experiment?
It is possible to safely interrupt the protocol after the fixation step. Thereto, remove the fixation solution and wash as suggested by the user manual, then the cells can be stored in buffer at 4° C. Alternatively, the experiment can also be safely interrupted after permeabilization, again as described above.
Please note: It is important to proceed with the experiment if the click cocktail for the detection of the EdU has been prepared already. -
Is antibody staining compatible and can be combined with the EdU?
Antibody staining is compatible and can be combied with EdU cell proliferation detection when antibody detection is done after the click detection step. Please be aware of the dye used for EdU detection. Check also the user manual for more information. The workflow is designed to allow multiplexing with surface and intracellular markers, provided staining is performed after the click reaction.
-
What is the recommended EdU incubation time?
Incubation time depends on the cell type and proliferation rate.
As a general guideline we recommend to use a maximum of 10 µM final EdU in the cell culture medium for incubations. Typical labeling ranges from 30 minutes to several hours. For slow-dividing cells, longer incubation (> 1 day) may be required and the concentration should be decreased to 1-5 µM.
-
How should I store the kit and its components?
Store the kit at 2–8 °C in the dark. Avoid repeated freeze-thaw cycles of fluorescent dyes to maintain performance.
-
Can I use this kit with fixed samples?
Yes, the kit is compatible with fixed and permeabilized cells. Follow the protocol for fixation and permeabilization steps to ensure optimal dye penetration.
-
Is the kit suitable for suspension cells as well as adherent cells?
Yes, the protocol supports both suspension and adherent cell types commonly analyzed by flow cytometry.
-
How do I choose the right fluorescent dye?
Select a dye based on your flow cytometer’s laser and filter settings. For example:
- 6-FAM for 488 nm (FITC channel)
- TAMRA for 555 nm
- Sulforhodamine for 594 nm
- Eterneon Red for 647 nm
-
How does the ClickTech EdU Flow Cytometry Kit compare to Thermo Fisher’s Click-iT™ Kit?
Both kits are based on the same EdU click chemistry technology for detecting DNA synthesis during cell proliferation. The main difference lies in pricing and flexibility.

