EdU Cell Proliferation Assay for Flow Cytometry
ClickTech EdU Cell Proliferation Kit for Flow Cytometry
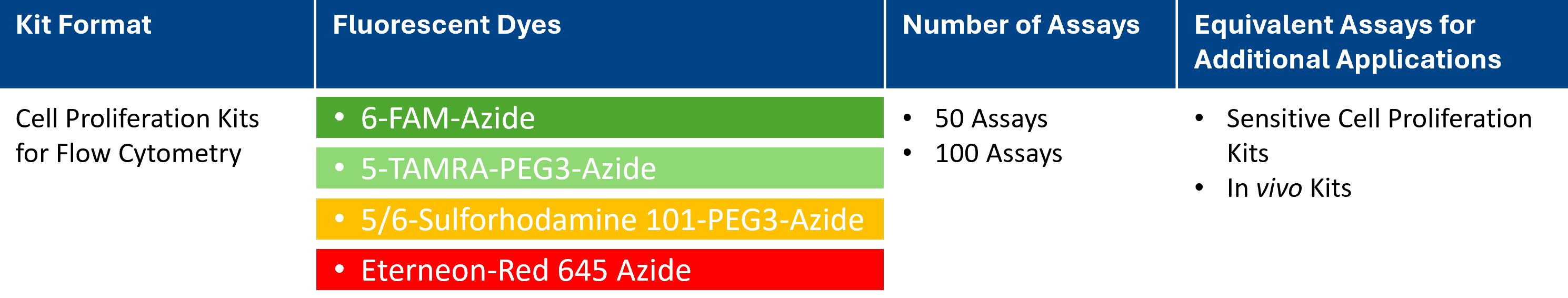
| Size | Catalog No. | Price |
|---|---|---|
| Dye 488 / 50 Assays | BCK-EdU488FC50 | € 495,00 |
| Dye 488 / 100 Assays | BCK-EdU488FC100 | € 770,00 |
| Dye 555 / 50 Assays | BCK-EdU555FC50 | € 495,00 |
| Dye 555 / 100 Assays | BCK-EdU555FC100 | € 770,00 |
| Dye 594 / 50 Assays | BCK-EdU594FC50 | € 495,00 |
| Dye 594 / 100 Assays | BCK-EdU594FC100 | € 770,00 |
| Dye 647 / 50 Assays | BCK-EdU647FC50 | € 495,00 |
| Dye 647 / 100 Assays | BCK-EdU647FC100 | € 770,00 |
-
Cell Analysis – Cell Proliferation Flow Cytometry Assays
Cell analysis allows scientists to gain unique insights into cell proliferation, viability, and toxicity, enabling the monitoring of cell response and cell health in cultures after treatment with various stimuli.
Detection of DNA replication is a powerful tool for studying proliferation of both cell populations and individual cells. DNA replication rates can be detected through the incorporation of a nucleoside analog such as BrdU or EdU into DNA. The detection of these two thymidine analogs differs significantly and has implications for test results, potential applications, and multiplexing capability. Compared to other cell proliferation methods, the EdU proliferation assay does not involve radioactive isotopes or antibodies to detect newly synthesized DNA.
The ClickTech EdU cell proliferation assay enables estimates of cell proliferation rates by determining the fraction of cells replicating their DNA.
The technology is based on the labeling of DNA with EdU (5-ethynyl-2′-deoxyuridine), an alkyne-modified nucleoside that is incorporated into the replicating DNA of living cells and its conjugation to a fluorochrome in situ.
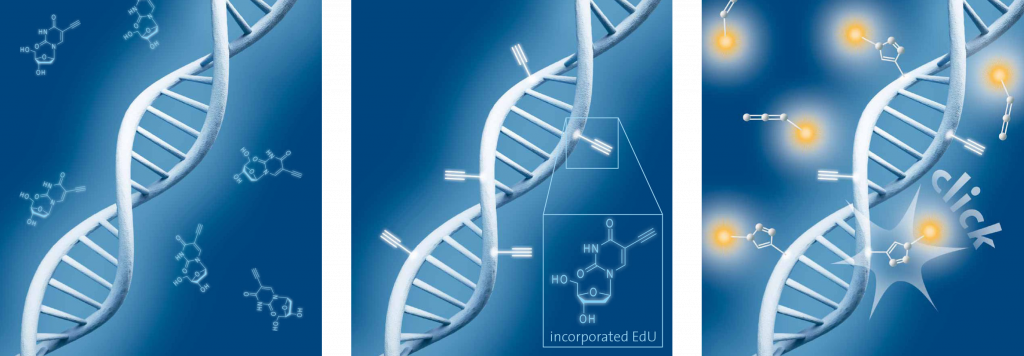
Schematic representation of EdU cell proliferation assays. A) Incubation of cells with EdU. B) EdU is incorporated during active DNA synthesis. C) Detection of cell proliferation via click chemistry, wherein the use of a range of different fluorescent dyes is possible.
Advantages:
- Increased selectivity and sensibility in comparison to BrdU
- Simple, fast and reliable assay
- High sensitivity paired with very low toxicity
- A cost-effective assay
- Multiplexing possibility
- higher preservation of morphological details relative to BrdU
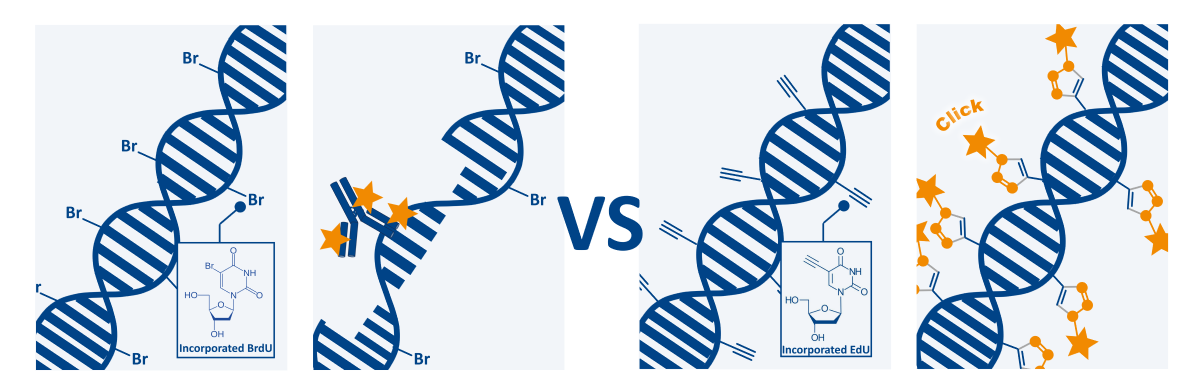
ClickTech EdU Cell Proliferation Assays for FC
The ClickTech EdU Cell Proliferation Kit for FC is a simple and stable assay perfectly suited for monitoring genotoxicity, anticancer drug evaluation or cell cycle analysis by directly measuring the proportion of cells that replicate their DNA. The principle of this procedure is similar to that of the BrdU assay, with the difference of the detection reaction. In summary, the thymidine analog, in this case the EdU (5-ethynyl-2′-deoxyuridine), is added to the cell culture media, taken up by the cells and incorporated into newly synthesized DNA in cell division. This nucleoside contains a reactive alkyne group (instead of the bromine atom in BrdU) and incorporated nucleotides can undergo a click reaction. After an incubation period that depends on the specific cell type, the cells are washed, fixed and permeabilized to allow the detection reagents to penetrate. The detection is then carried out via the copper-catalyzed click reaction (also known as Azide-Akyne Cyclo-addition or CuAAC). Click reactions show high quantitative yields, run under mild reaction conditions and are easy to perform. In order to start the detection reaction, a click cocktail, containing buffer, catalytic reagents and a fluorescent dye, is added to the cells . The dye contains an azide group that reacts with the alkyne group of the incorporated EdU. Only cells that actively replicate their DNA during the EdU pulse can be detected by this method.
Workflow
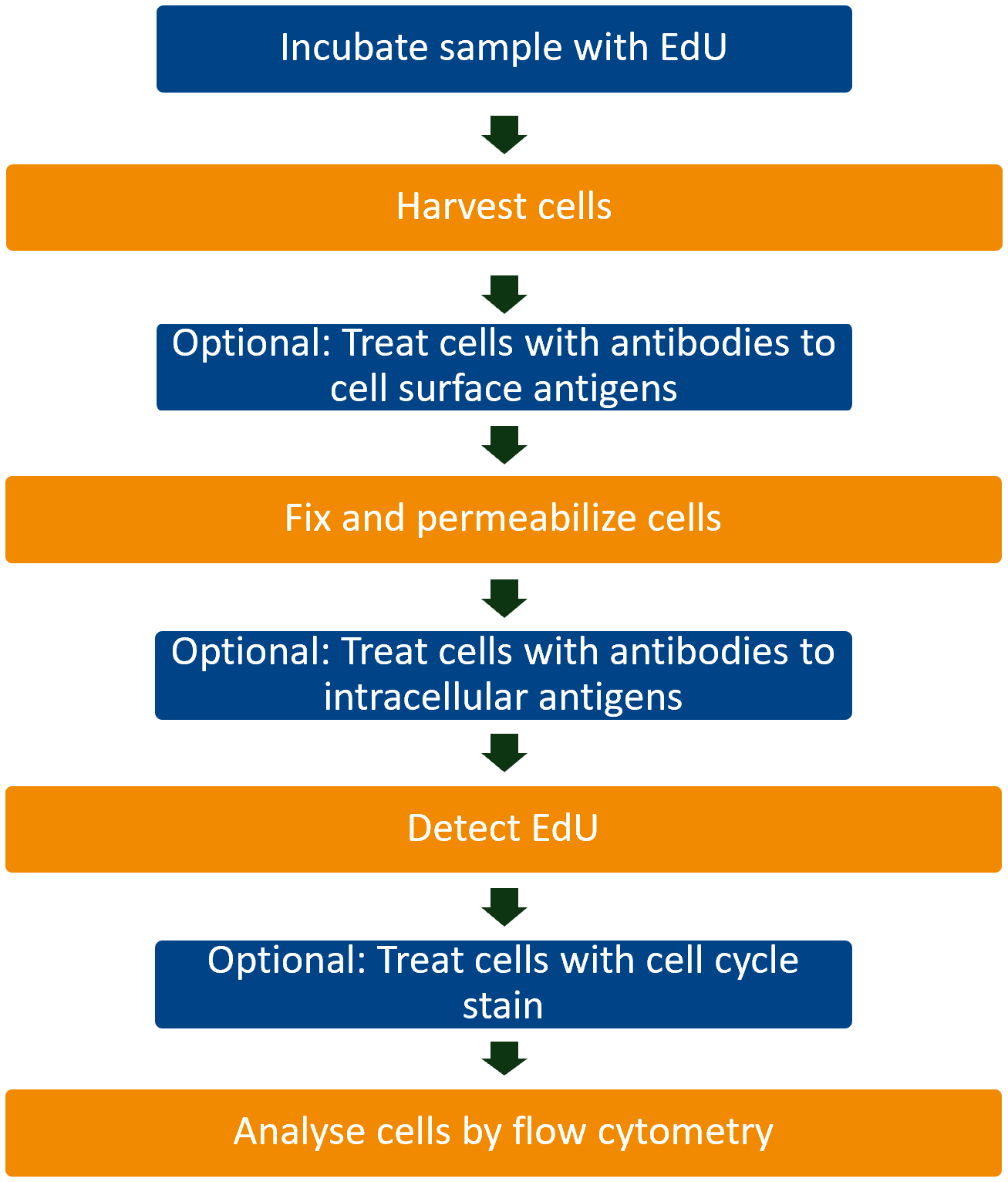
EdU Detection
Functional alkyne and azide groups do not occur in natural cell systems and do not react without catalytic reagents. This ensures that the EdU cell proliferation method, based on click chemistry, is highly selective, sensitive and reproducible. This technology allows an almost unlimited choice of different and easy to synthesize fluorescent dyes and thus offers maximum flexibility in terms of “detection colors” and analytical devices. Cell proliferation can be detected by fluorescence microscopy, flow cytometry and fluorescence plate reader. baseclick currently offers assays for these three analytical methods and up to four standard dyes (488, 555, 594, 647 nm).
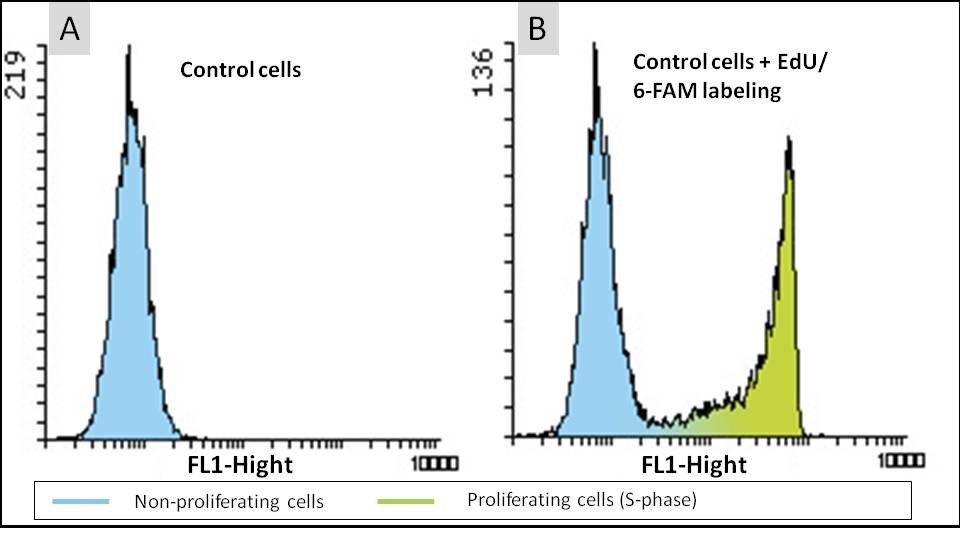
Cell proliferation assay with the EdU Cell Proliferation Kit for Flow Cytometry (BCK-EdU488FC). HeLa cells were either untreated (A) or incubated with 10 µM EdU for 2 hours (B). The click reaction was performed using 6-FAM Azide and fluorescence intensity of 10.000 cells was measured by flow cytometry. The results are presented in form of histograms, showing the cell number in the y-axis and the FL1-Fluorescence in the x-axis.
Product Overview
LITERATURE
Cytolethal Distending Toxin Promotes Replicative Stress Leading to Genetic Instability Transmitted to Daughter Cells, W. Tremblay et al., 2021, Frontiers in Cell and Developmental Biology, Vol. 9, p. 656795.
https://doi.org/10.3389/fcell.2021.656795
Innovative DNA-Targeted Metallo-prodrug Strategy Combining Histone Deacetylase Inhibition with Oxidative Stress, T. McGivern et al., 2018, Mol. Pharmaceutics, Vol.15, p. 5058-5071.
https://doi.org/10.1021/acs.molpharmaceut.8b00652
YB-1 Expression and Phosphorylation Regulate Tumorigenicity and Invasiveness in Melanoma by Influencing EMT, C. Kosnopfel et al., 2018, Mol Cancer Res., Vol. 16, p. 1149-1160.
https://doi.org/10.1158/1541-7786.MCR-17-0528
Chronic exposure to Cytolethal Distending Toxin (CDT) promotes a cGAS-dependent type I interferon response, B. J. Pons et al., 2021, Cellular and Molecular Life Sciences, Vol. 78, p. 6319–6335.
FAQ
-
What type of cells can incorporate EdU?
The EdU cell proliferation assay has been applied to many different cell types and organisms from prokaryotic to eukaryotic. Cell lines such as E. coli, HeLa, HEK, MOLM are arguably among the most routine applications, but also animals, like mouse, rat, the nematode C. elegans, crickets (Gryllus bimaculatus), chicken (Gallus domesticus) and zebra fish (Danio rerio) or even plants (e.g. Arabidopsis thaliana) can be applied.
Cells that possess a pyrimidine pathway that can phosphorylate EdU to the corresponding triphosphate, which is then accepted by the host DNA polymerase for incorporation into DNA during replication. -
Can I perform EdU cell proliferation detection on living cells?
EdU is incorporated into living cells, but the detection reaction must be performed on fixed and permeabilized samples.
-
How does EdU labeling compare to BrdU or the 3H-thymidine incorporation assay?
All three methods enable to determine cell proliferation directly by incorporation of a metabolite analogue and subsequent detection. The 3H-thymidine incorporation assay is very sensitive, but the radioactive compound requires specialized equipment and dedicated lab space for handling. EdU and BrdU assays are non-radioactive alternatives with decreased risk for health and environment. Compared to the BrdU incorporation assay the EdU assay is more sensitive, requires less handling time and needs no harsh DNA denaturing conditions for detection. Therefore, the EdU cell proliferation is also compatible with multiplexing.
-
Can I combine DAPI staining and EdU detection?
Yes, this is feasible. Please note that DAPI staining should be done after the click detection step. Alternatively, SYBR Green DNA staining can be used. But, please note that SYBR green should not be used with dyes of 488 nm wavelengths.
-
When can I safely interrupt the experiment?
It is possible to safely interrupt the protocol after the fixation step. Thereto, remove the fixation solution and wash as suggested by the user manual, then the cells can be stored in buffer at 4° C. Alternatively, the experiment can also be safely interrupted after permeabilization, again as described above.
Please note: It is important to proceed with the experiment if the click cocktail for the detection of the EdU has been prepared already. -
Is antibody staining compatible with the EdU?
Antibody staining is compatible with EdU cell proliferation detection when antibody detection is done after the click detection step. Please be aware of the dye used for EdU detection. Check also the user manual for more information.
-
How to determine the EdU incubation time?
The EdU incubation time depends on the cell type or organism, the applied EdU concentration and the experimental design. For a start it is advisable to refer to a literature protocol (which is close to your experimental setup) and to test the conditions with a low number of samples. As a general guideline we recommend to use a maximum of 10 µM final EdU in the cell culture medium for incubations. For longer incubation (> 1 day) the concentration should be decreased to 1-5 µM.
-
-
Shelf Life
12 months unopened after receipt
-
Storage Conditions
2-8 °C
-
Physical State
kit system made of different components
-
CAS Number
n.a.
-
Excitation (max)
Dye 488: 496 nm | Dye 555: 546 nm | Dye 594: 584 nm | Dye 647: 643 nm
-
Emission (max)
Dye 488: 516 nm | Dye 555: 579 nm | Dye 594: 603 nm | Dye 647: 662 nm
-
Ɛ (max)
Dye 488: 83.000 cm-1M-1 | Dye 555: 91.000 cm-1M-1 | Dye 594: 110.000 cm-1M-1 | Dye 647: 250.000 cm-1M-1
-
Preparation/Handling
please see user manual of the kit
-
Shelf Life

