
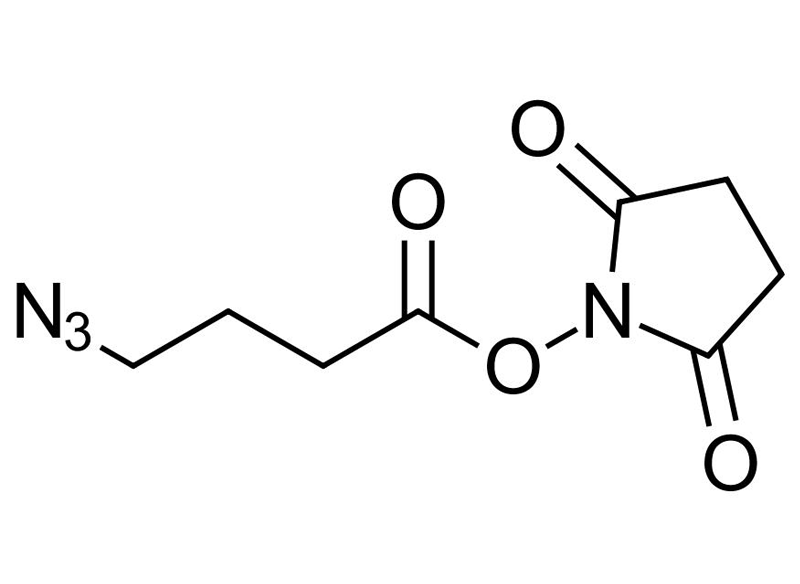
NHS-C3-Azide
Bifunctional linker
| Size | Catalog No. | Price |
|---|---|---|
| 5 mg | BCL-014-5 | € 40,00 |
| 10 mg | BCL-014-10 | € 65,00 |
Chemical Properties
-
Molecular Formula
C8H10N4O4
-
Shelf Life
12 months unopened after receipt
-
Storage Conditions
-20 °C, dry, inert gas
-
Molecular Weight
226.19 g/mol
-
Purity
≥ 95% (NMR)
-
Physical State
White to off-white solid
-
CAS Number
943858-70-6
-
Additional name
gamma-Azidobutyric acid oxysuccinimide ester; γ-Azidobutyric acid NHS; 2,5-dioxopyrrolidin-1-yl 4-azidobutanoate; Azidobutyric NHS ester
-
Solubility
DMSO, DMF, DCM
-
Preparation/Handling
For a 10 mM solution add 442 μL to 1 mg.
Product Information
Azidobutyric NHS Ester for Bioorthogonal Conjugation and Click Chemistry
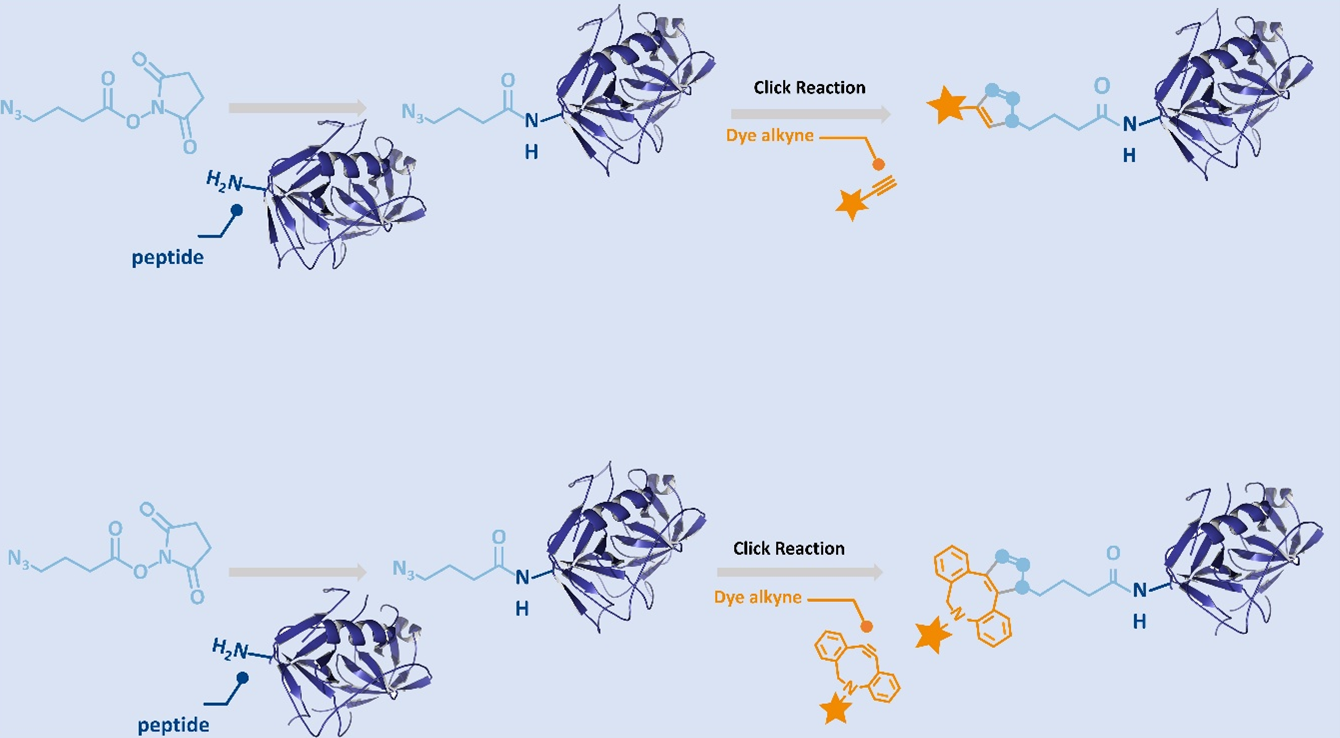
NHS-C3-Azide, also known as Azidobutyric NHS ester, is a hetero-bifunctional crosslinking reagent that combines an N-hydroxysuccinimide (NHS) ester and an azido (-N₃) group. This dual functionality makes it an ideal tool for bioorthogonal labeling, surface functionalization, and controlled biomolecular modification.
The NHS ester component enables selective and efficient coupling to primary amines in biomolecules such as proteins, peptides, antibodies, and amine-modified oligonucleotides. Meanwhile, the azide group participates in copper-catalyzed (CuAAC) or copper-free (SPAAC) click reactions, forming stable triazole linkages with alkynes or strained cyclooctynes like DBCO or BCN.
Traditional conjugation chemistries often suffer from poor selectivity, harsh conditions, or incompatibility with delicate biomolecules.
NHS-C3-Azide provides a highly selective, biocompatible, and stable platform for orthogonal bioconjugation, enabling precise molecular engineering in pharmaceutical development and advanced biotechnology research, without compromising biomolecule function or structure. The main advantages of NHS-C3-Azide are:
- Dual Reactivity: Ideal for introducing azides into amine-modified oligos after solid-phase synthesis, avoiding degradation risks from azides during P(III)-based phosphoramidite chemistry.
- Fast & Efficient Click Reactions: Forms stable triazole bonds with alkynes and cyclooctynes (e.g., DBCO, BCN)
This reagent is used for:
- Biomolecule labeling (proteins, peptides, nucleic acids)
- Targeted drug delivery conjugates
- Surface and hydrogel modifications
- Antibody and aptamer functionalization
LITERATURE
Microstructured click hydrogels for cell contact guidance in 3D, M.I. Neves et al., 2023, Materials Today Bio, Vol.19, p.100604.
https://doi.org/10.1016/j.mtbio.2023.100604.
Real-time analysis of quantum dot labeled single porcine epidemic diarrhea virus moving along the microtubules using single particle tracking, W. Hou et al., 2019, Scientific Reports, Vol. 9, p. 1307.
https://doi.org/10.1038/s41598-018-37789-9
Integration of a field effect transistor-based aptasensor under a hydrophobic membrane for bioelectronic nose applications, A.E. Kuznetsov et al., 2019, Biosensors and Bioelectronics, Vol. 129, p. 29-35.
https://doi.org/10.1016/j.bios.2019.01.013
Enterotoxigenic Escherichia coli–blood group A interactions intensify diarrheal severity, P. Kumar et al., 2018, J. Clin. Investig., Vol. 128(8), p. 3298-3311.
https://doi.org/10.1172/JCI97659DS1
Amplified Detection of the Aptamer–Vanillin Complex with the Use of Bsm DNA Polymerase, M. Andrianova et al., 2018, Sensores, Vol. 18, p. 49.
https://doi.org/10.3390/s18010049
Aptamer based vanillin sensor using an ion-sensitive field-effect transistor, A. Kuznetsov et al., 2018, Microchimica Acta, Vol. 185(1), p. 3.
https://doi.org/10.1007/s00604-017-2586-4
Tandem Oligonucleotide Probe Annealing and Elongation To Discriminate Viral Sequence, M. Taskova et al., 2017, Analytical Chemistry, Vol. 89(8), p. 4363–4366.
https://doi.org/10.1021/acs.analchem.7b00646
Gas phase click chemistry via ion/ion reactions, J. Bu et al., 2015, Int J Mass Spectrom, Vol. 309, p. 118-123.
https://doi.org/10.1016/j.ijms.2015.05.010
Two-step labeling of Staphylococcus aureus with Lysostaphin-Azide and DIBO-Alexa using click chemistry, I. Potapova et al., 2013, Journal of Microbiological Methods, Vol. 92(1), p. 90-98.

