
Poly(A)-ClickSeq Library Prep Kit
Selective 3′-end mRNA sequencing using click chemistry

| Size | Catalog No. | Price |
|---|---|---|
| 12 rxn | BCK-PACSeq | € 396,00 |
Chemical Properties
-
Shelf Life
12 months unopened after receipt
-
Storage Conditions
– 20 °C
-
Physical State
kit system made of different components
-
CAS Number
n.a.
-
Preparation/Handling
please see user manual of the kit
Product Information
Poly(A)-ClickSeq (PACSeq) is a targeted RNA sequencing technique that focuses specifically on sequencing the 3′ ends of polyadenylated RNA, such as eukaryotic mRNAs. As a specialized form of RNA-Seq, PACSeq offers a streamlined and cost-effective approach for profiling poly(A) sites and alternative polyadenylation, while maintaining key advantages of traditional RNA-Seq methods.
Unlike conventional whole-transcript sequencing, PACSeq avoids unnecessary fragmentation and emphasizes the biologically important 3′ UTR region.
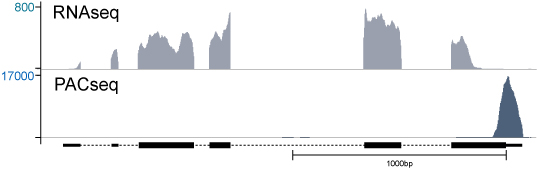
Comparison of RNAseq technique with PACseq technique.
The technology behind PACSeq:
PACSeq utilizes a modified reverse transcription reaction that includes three chain-terminating nucleotides next to the natural ones:
No thymidine analog is used, so reverse transcription only is able to be terminated after the last adenine (A) of the poly(A) tail, ensuring selective capture of the 3′ UTR region. For faster chain termination a different ratio of natural to terminating azido nucleotides is used in PACseq in full sequence reads. This new approach enables both poly(A) site mapping and gene expression profiling in one workflow.
Advantages of PACSeq Over Traditional RNA-Seq
- No fragmentation required
- No enzymatic ligation, reducing artifacts and recombination
- Works with fragmented or degraded RNA
- Stranded technique provides strand-of-origin information
- Ideal for rare recombination or chimeric event detection
- Low input required as little as 10 ng total RNA
- Rapid protocol, libraries ready in ~6 hours
- Unique Molecular Identifiers (UMIs) are available
PACSeq maintains the robust performance of RNA-Seq while simplifying the workflow, lowering costs, and enriching for biologically relevant 3′ ends.
Applications
- Mapping of polyadenylation sites
- Studies of alternative polyadenylation (APA)
- Gene expression profiling with 3′ bias
- Quantifying transcript isoforms and post-transcriptional regulation
- Suitable for degraded RNA samples (e.g. FFPE)
Kit Components and Requirements
The Poly(A)-ClickSeq Library Prep Kit includes all core reagents required for PACSeq library generation except PCR primers.
If needed, a set of 12 compatible i7 index primers can be purchased separately here. Alternatively, users may provide their own primers if already validated for Illumina sequencing workflows.
LITERATURE
For a detailed list of publications utilizing PACseq technology please visit:
https://www.clickseqtechnologies.com/resources/polya-clickseq-papers
FAQ
-
What is Poly(A)-ClickSeq?
Poly(A)-ClickSeq is a specific variant of ClickSeq that uses an Oligo-dT primer in the Reverse transcription step. cDNA fragments are thus focused to the 3’ ends of polyadenylated RNAs, such as metazoan mRNAs and viral genomic RNAs. As such, Poly(A)-ClickSeq provides a powerful platform for transcriptomics and differential gene expression analysis. Since only one NGS read is obtained per molecule of mRNA in a sample, the bioinformatics procedures are greatly simplied and the amount of sequence depth is greatly reduced (~10M reads required vs 100M per sample for a typical transcriptomics study). Additionally, Poly(A)-ClickSeq can be used to precisely map the junction of a poly(A)-tail within the 3’UTR of a mRNA. As such, it is an excellent tool for studying alternative polyadenylation and alternative 3’ exon usage.
Poly(A)-ClickSeq retains all the advantages of ClickSeq, such as the removal of fragmentation steps and a simplified workflow. In addition, Poly(A)-ClickSeq does not require any ribo-depletion or mRNA enrichment prior to the library prep, since the Oligo-dT primer automatically enriches for mRNAs and other species of interest. In some cases, Poly(A)-ClickSeq can even be performed directly on cells, without RNA extraction.
-
Where can I find a protocol for Poly(A)-ClickSeq?
Our protocol can be found here:
https://www.protocols.io/view/poly-a-clickseq-poly-a-primed-protocol-with-single-n2bvjnb5xgk5/v1
-
Is Poly(A)-ClickSeq published?
Yes. Poly(A)-ClickSeq was originally published in 2017 by Routh et al in Nucleic Acids Research (NAR):
-
What do I need in addition to the kit to make a ClickSeq library?
Required third-party reagents include:
- SuperScript III™ Reverse Transcriptase, 200U/µL (Invitrogen; 18080-093 or 18080-044)
- ClickSeq Library Prep Kit Index Primers (baseclick, BCK-RNAseq-IP)
- [optional] RNaseOUT™ Recombinant Ribonuclease Inhibitor, 40U/µL (Invitrogen; 10777)
- OneTaq® 2X Master Mix with Standard Buffer (NEB; M0482S or M0482L) (Note: you must use OneTaq for this step, this enzyme cannot be substituted for a different PCR enzyme)
- [optional] RNase H, 5000 units/mL (NEB; M0297S or M0297L)
- SPRIselect (Beckman Coulter; B23317) or equivalent DNA/RNA Purification Beads (also known as SPRI beads)
- Nuclease free water
- 80% ethanol (made fresh)
-
Where can I find more information?
Please see an extensive set of FAQs:

