
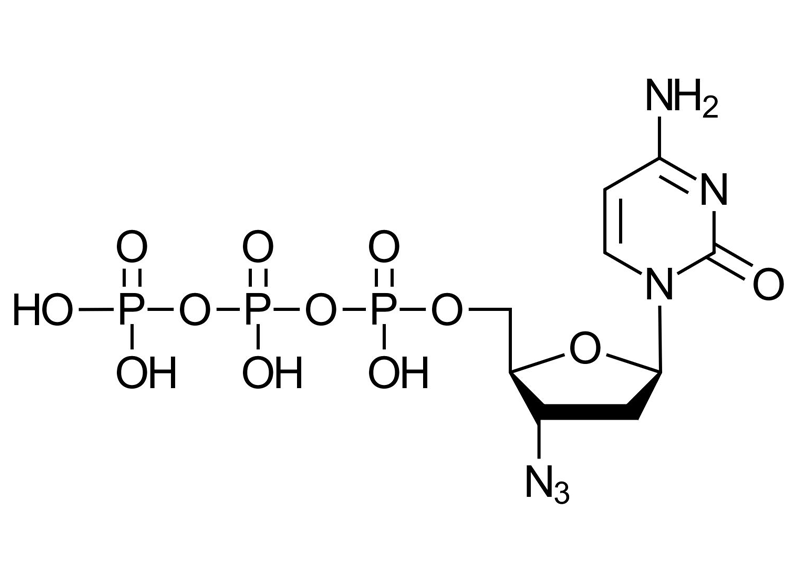
3′-Azido-2′,3′-ddCTP
Triphosphate for modifying of 3'END RNA or ssDNA
| Size | Catalog No. | Price |
|---|---|---|
| 1 µmol | BCT-26-S | € 250,00 |
| 5 µmol | BCT-26-L | € 850,00 |
Chemical Properties
-
Molecular Formula
C9H15N6O12P3
-
Shelf Life
12 months unopened after receipt
-
Storage Conditions
-20 °C
-
Molecular Weight
492.17 g/mol
-
Purity
≥ 98% (HPLC)
-
Physical State
100 mM clear aquaeous solution; colorless
-
CAS Number
92562-77-1 (free acid)
-
Absorption (max)
λmax = 271 nm
-
Ɛ (max)
8,900 cm-1M-1
Product Information
A nucleotide analog for selective 3`end labeling of nucleic acids via click chemistry or Staudinger ligation
Overview and Molecular Structure
3′-Azido-2′,3′-ddCTP is a modified nucleotide analog of cytidine triphosphate (CTP), featuring a 3`azido group at the 3′ position instead of the natural 3`OH group. As a dideoxy nucleotide, the lack of 2`OH group, making it a chain terminating nucleotide during polymerase reactions.
Unlike internal labeling nucleotides (e.g., EdUTP or EUTP) 3′-Azido-2′,3′-ddCTP specifically designed for post-synthetic 3′-end modification. It is incorporated enzymatically using terminal deoxynucleotidyl transferase (TdT) or poly(A) polymerase, ensuring single-site labeling at the 3′ end.
Chemical functionality of 3′-Azido-2′,3′-ddCTP:
The azido group is a biorthogonal functionality, meaning it does not interfere with natural biomolecules. It reacts selectively with:
- Alkynes → forming triazoles via CuAAC (copper-catalyzed) or SPAAC (copper-free, using strained alkynes like DBCO)
- Phosphines → forming amides via Staudinger ligation
Both reactions meet the criteria for click chemistry, and the development of azide–alkyne cycloaddition was recognized with the 2022 Nobel Prize in Chemistry.
Typical conjugation partners include fluorescent dyes, biotin, linkers, and advanced ligands such as sugars for targeted delivery.
Application areas for 3′-Azido-2′,3′-ddCTP
- 3′-End Labeling of Nucleic Acids: Incorporation into ssDNA or mRNA followed by click conjugation for imaging or functionalization.
- Reverse Transcription and cDNA Synthesis: Acts as a chain terminator during reverse transcription, enabling controlled fragment generation for sequencing workflows.
- Next-Generation Sequencing (NGS): Used in ClickSeq kits as a chain terminator, 3′-Azido-2′,3′-ddCTP generates azido-modified fragments that enable primer or adapter conjugation via click chemistry, eliminating the need for enzymatic ligation. Like Sanger sequencing, which relies on ddNTPs to stop elongation for sequence determination, ClickSeq applies the same principle using azido-modified dideoxynucleotides. These nucleotides terminate synthesis at defined positions and introduce an azide group, allowing click chemistry-based adapter or primer conjugation without enzymatic ligation. This approach combines the precision of Sanger termination with the flexibility of modern biorthogonal chemistry, reducing artifacts such as chimeras, recombination, and ligation bias, while preserving library complexity and improving reproducibility in NGS workflows.
- Advanced Bioconjugation: Enables attachment of targeting moieties for active transport into cells.
Advantages
- Site-specific labeling at the 3′end
- Guaranteed single incorporation due to chain termination
- Compatible with copper-free click chemistry for sensitive biomolecules
- Ideal for sequencing and controlled fragment generation.
LITERATURE
Site-specific terminal and internal labeling of RNA by poly(A) polymerase tailing and copper-catalyzed or copper-free strain-promoted click chemistry, M. L. Winz et al., 2012, Nucleic Acids Res., Vol. 40, p. 1–13.
https://doi.org/10.1093/nar/gks062
Chemoenzymatic Preparation of Functional Click-Labeled Messenger RNA, S. Croce et al., 2020, ChemBioChem, Vol. 21, p. 1641-1646.
https://doi.org/10.1002/cbic.201900718
Application and design considerations for 3′-end sequencing using click-chemistry, M. K. Jensen et al., 2021, Methods in Enzymology, Vol. 655, p. 1-23.
https://doi.org/10.1016/bs.mie.2021.03.012
Trimannose-coupled antimiR-21 for macrophage-targeted inhalation treatment of acute inflammatory lung damage, C. Beck et al., 2023, Nature Communications, Vol. 14, 4564.
https://www.nature.com/articles/s41467-023-40185-1
FAQ
-
What is the primary application of 3′-Azido-2′,3′-ddCTP?
It is a nucleotide analog designed for selective 3′-end labeling of nucleic acids via click chemistry or Staudinger ligation. This enables site-specific conjugation of functional groups such as fluorophores, biotin, or affinity tags.
-
Which enzymes can incorporate 3′-Azido-2′,3′-ddCTP?
It is compatible with DNA and RNA polymerases that tolerate modified nucleotides. For optimal results, use polymerases validated for incorporation of dideoxynucleotides in chain termination assays.
-
Can this analog be used in sequencing workflows?
Yes. It is suitable for applications requiring controlled chain termination and clickable handles for downstream conjugation, such as Sanger sequencing or specialized NGS library preparation.
-
What click chemistry reactions are supported?
The azide group enables bioorthogonal conjugation via Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC) and Staudinger ligation under mild conditions.
-
Is it compatible with live-cell labeling?
No. This analog is intended for in vitro enzymatic reactions and post-synthetic labeling of nucleic acids, not for direct incorporation in live cells.
-
How should the product be stored?
Store at −20 °C in a dry environment. Avoid repeated freeze-thaw cycles to maintain integrity.
-
What are typical downstream applications?
Fluorescent labeling for imaging, Affinity tagging for pull-down assays, Preparation of modified oligonucleotides for structural studies

