
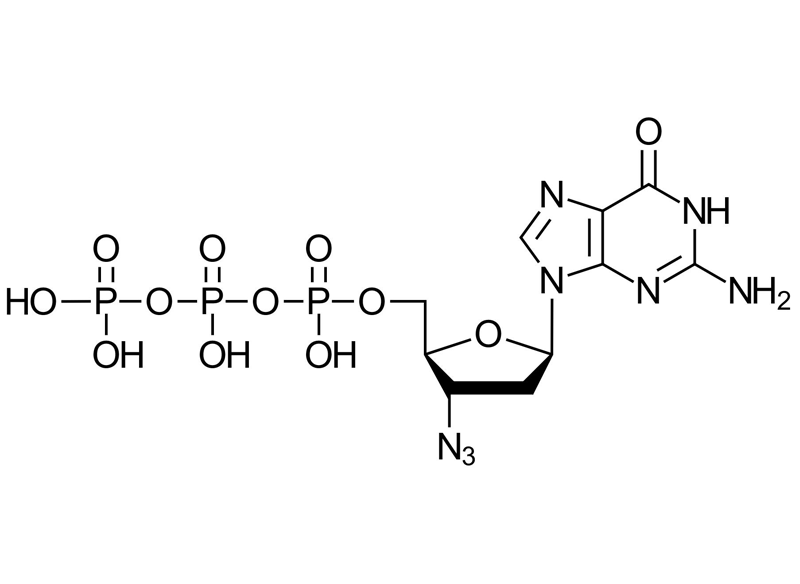
3′-Azido-2′,3′-ddGTP
Triphosphate for modifying of 3'END RNA or ssDNA
| Size | Catalog No. | Price |
|---|---|---|
| 1 µmol | BCT-27-S | € 250,00 |
| 5 µmol | BCT-27-L | € 850,00 |
Chemical Properties
-
Molecular Formula
C10H15N8O12P3
-
Shelf Life
12 months unopened after receipt
-
Storage Conditions
-20 °C
-
Molecular Weight
532.20 g/mol
-
Purity
≥ 98% (HPLC)
-
Physical State
100 mM clear aquaeous solution; colorless
-
CAS Number
94059-38-8 (free acid)
-
Absorption (max)
λmax = 252 nm
-
Ɛ (max)
13,700 cm-1M-1
Product Information
A Nucleotide Analogue for the Selective 3´End Labeling of Nucleic Acids via Click Chemistry or Staudinger Ligation
Molecular Structure and Overview
3′-Azido-2′,3′-ddGTP is a chemically modified analogue of guanosine triphosphate (GTP) featuring an azido (N₃) group in place of the 3′OH group of the sugar moiety. This modification enables highly efficient labelling through click chemistry reactions, including:
- copper-catalyzed alkyne-azide cycloaddition (CuAAC)
- copper-free strain-promoted alkyne-azide cycloaddition (SPAAC).
The absence of the 3′-OH group makes 3′-Azido-2′,3′-ddGTP a chain-terminating nucleotide in polymerization reactions such as IVT. Unlike other modified NTPs (e.g., EUTP or EdUTP), it cannot be incorporated internally during nucleic acid synthesis. Instead, it is ideal for post-synthetic, 3′-end-specific introduction of a single azido-modified guanosine using enzymes such as T7 polymerase or terminal deoxynucleotidyl transferase (TdT).
Detailed protocols for fluorescent dye, biotin, linker, or ligand conjugation via click chemistry are available from baseclick.
Role in Sequencing and Research Applications
The chain-terminating property of 3′-Azido-2′,3′-ddGTP is utilized in baseclick’s ClickSeq NGS sequencing kits to generate azido-modified short nucleic acid fragments during reverse transcription. Similar in our ClickTech Long-Read Library Prep Kits cDNA fragments are selectively elongated by 3′-Azido-2′,3′-ddGTP incorporation to be accessable for primer elongation by Click Chemistry.
Azide Functionality and Biorthogonal Ligation Reactions:
The azido group is bioorthogonal, reacting only with:
- Alkynes: Forms triazoles under Cu(I) catalysis or with strained alkynes (e.g., DBCO, BCN) without a catalyst.
- Phosphines: Forms amides via Staudinger ligation.
Both reactions meet the criteria of click chemistry, and the development of azide–alkyne cycloaddition was recognized with the 2022 Nobel Prize in Chemistry. Typical conjugation partners include fluorescent dyes, biotin, linkers, and advanced ligands such as sugars for targeted delivery.
Applications
- 3′-Azido-2′,3′-ddGTP is primarily used for site-specific 3′-end labeling of nucleic acids such as ssDNA or mRNA. After enzymatic incorporation, the azido group enables versatile chemical modifications via click chemistry for imaging, affinity tagging, or functionalization.
Beyond labeling, its chain-terminating property makes it valuable in sequencing workflows:
- Click Chemistry Applications: Enables conjugation with fluorescent dyes, biotins, linkers, or cell-targeting ligands.
- Telomerase inhibition: 3′-Azido-2′,3′-ddGTP can inhibit telomerase activity without affecting DNA polymerases. This enables their potential application in anticancer research by reducing telomerase activity, limiting cell division, and promoting apoptosis in cancer cells.
Summary of Common Uses
- mRNA and ssDNA labeling via controlled enzymatic incorporation.
- Chain termination in NGS workflows for improved sequencing accuracy.
LITERATURE
Site-specific terminal and internal labeling of RNA by poly(A) polymerase tailing and copper-catalyzed or copper-free strain-promoted click chemistry, M. L. Winz et al., 2012, Nucleic Acids Res., Vol. 40, p. 1–13.
https://doi.org/10.1093/nar/gks062
Chemoenzymatic Preparation of Functional Click-Labeled Messenger RNA, S. Croce et al., 2020, ChemBioChem, Vol. 21, p. 1641-1646.
https://doi.org/10.1002/cbic.201900718
Application and design considerations for 3′-end sequencing using click-chemistry, M. K. Jensen et al., 2021, Methods in Enzymology, Vol. 655, p. 1-23.
https://doi.org/10.1016/bs.mie.2021.03.012
Nanopore ReCappable sequencing maps SARS-CoV-2 5′ capping sites and provides new insights into the structure of sgRNAs, C. Ugolini et al., 2022, Nucleic Acids Research, Vol 50(6), p. 3475–3489.
https://doi.org/10.1093/nar/gkac144
Identification of high-confidence human poly(A) RNA isoform scaffolds using nanopore sequencing, L. Mulroney et al., 2022, RNA, Vol. 28(2), p. 162-176.
https://doi.org/10.1261/rna.078703.121
FAQ
-
What is 3′-Azido-2′,3′-ddGTP?
It is a chemically modified guanosine triphosphate analogue featuring an azido (N₃) group instead of the 3′-OH group, enabling selective 3′-end labeling of nucleic acids via click chemistry.
-
How does it differ from other modified nucleotides like EUTP or EdUTP?
Unlike EUTP or EdUTP, 3′-Azido-2′,3′-ddGTP cannot be incorporated internally during nucleic acid synthesis. It acts as a chain terminator and is ideal for post-synthetic 3′-end labeling.
-
Which enzymes can incorporate 3′-Azido-2′,3′-ddGTP?
Commonly used enzymes include:
T7 RNA polymerase
Terminal deoxynucleotidyl transferase (TdT)
-
What click chemistry reactions are compatible?
CuAAC (Copper-catalyzed alkyne–azide cycloaddition)
SPAAC (Copper-free strain-promoted alkyne–azide cycloaddition)
Staudinger ligation
-
What are the main applications?
3′-end labeling of ssDNA and mRNA for imaging or functionalization
Chain termination in NGS workflows (e.g., ClickSeq kits)
Primer conjugation without enzymatic ligation
Potential telomerase inhibition for anticancer research
-
Can it be used for internal labeling of nucleic acids?
No. Due to the absence of the 3′-OH group, it cannot be incorporated internally during polymerization.
-
Is it used in sequencing kits?
Yes. It is a key component in baseclick’s NGS kits.
-
Does it affect DNA polymerases?
No. Studies indicate it inhibits telomerase activity but does not interfere with DNA polymerases.

