
RNA-ClickSeq Library Prep Kit
Fragmentation-free RNA library preparation for NGS

| Size | Catalog No. | Price |
|---|---|---|
| 12 rxn | BCK-RNASeq | € 396,00 |
Chemical Properties
-
Shelf Life
12 months unopened after receipt
-
Storage Conditions
– 20 °C
-
Physical State
kit system made of different components
-
CAS Number
n.a.
-
Preparation/Handling
please see user manual of the kit
Product Information
Fragmentation-free RNA library preparation for NGS
Introduction to ClickSeq Technology
ClickSeq is a novel approach to Next-Generation Sequencing (NGS) library preparation that replaces enzymatic ligation with a bioorthogonal chemical reaction, specifically, copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC). This highly efficient and specific reaction is used to attach sequencing adapters to newly synthesized cDNA molecules.
ClickSeq is designed to eliminate common bottlenecks and artifacts in standard RNA-sequencing workflows, such as sequence chimeras, recombination, fragmentation bias, and ligation inefficiencies, all while preserving library complexity and improving reproducibility.
RNA ClickSeq Library Preparation Kit
The RNA ClickSeq Kit is a complete solution for preparing high-quality NGS libraries directly from RNA via a fragmentation-free, ligation-free process.
The protocol begins with a reverse transcription (RT) step using a mixture of standard dNTPs and 3′-azido-2′,3′-dideoxynucleotides (AzNTPs). These modified nucleotides stochastically terminate the cDNA, blocking the 3′ end with an azide group. Instead of relying on enzyme-based adapter ligation, the cDNA fragments are subsequently click-ligated to alkyne-functionalized sequencing adapters.
This click-ligation step forms a stable triazole linkage, which is biocompatible with downstream PCR amplification using standard polymerases.
Problems with traditional RNA-Seq Workflows
Current RNA-seq methods are built on workflows that rely on enzymatic steps for adapter ligation and RNA fragmentation. These steps present multiple limitations:
- Enzymatic ligation introduces recombination artifacts, chimeras, and sequence biases
- Fragmentation steps require optimization and can damage RNA integrity
- Workflow complexity increases time, cost, and risk of failure
- High sample input is often required for sufficient library quality
- Chimera formation leads to data misinterpretation and false discovery
The advantages of Clickseq RNA library preparation assay
The ClickSeq protocol replaces this step by utilizing highly efficient and high yielding Cu(I) catalyzed alkyne azide cycloaddition (CuAAC) for ligation. Therefore, the incorporated 3`-Azido stop nucleotides are click ligated by CuAAC with an alkyne containing adapter Oligo. This results in a ssDNA molecule with a triazole linked DNA backbone, which is biocompatible for PCR amplification. For the defined 3`end by the stop azido nucleotide, no dual primers for both directions are needed.
These facts in combination with the usage of click chemistry instead of enzymatic ligation also significantly reduce the costs for preparing your NGS library. Additionally, ClickSeq with 6-8 h worktime is a lot faster than comparable procedures and due to the randomized stop nucleotides, no fragmentation of DNA is necessary.
Workflow of this Next Generation Sequencing (NGS) Library Prep Kit:
- Reverse Transcription: Incorporation of 3′-azido nucleotides into the cDNA.
- Click-Ligation: Connection of cDNA fragments with alkyne-functionalized sequencing adapters.
- PCR Amplification: Amplification of cDNA-adapter fragments to create the NGS library.
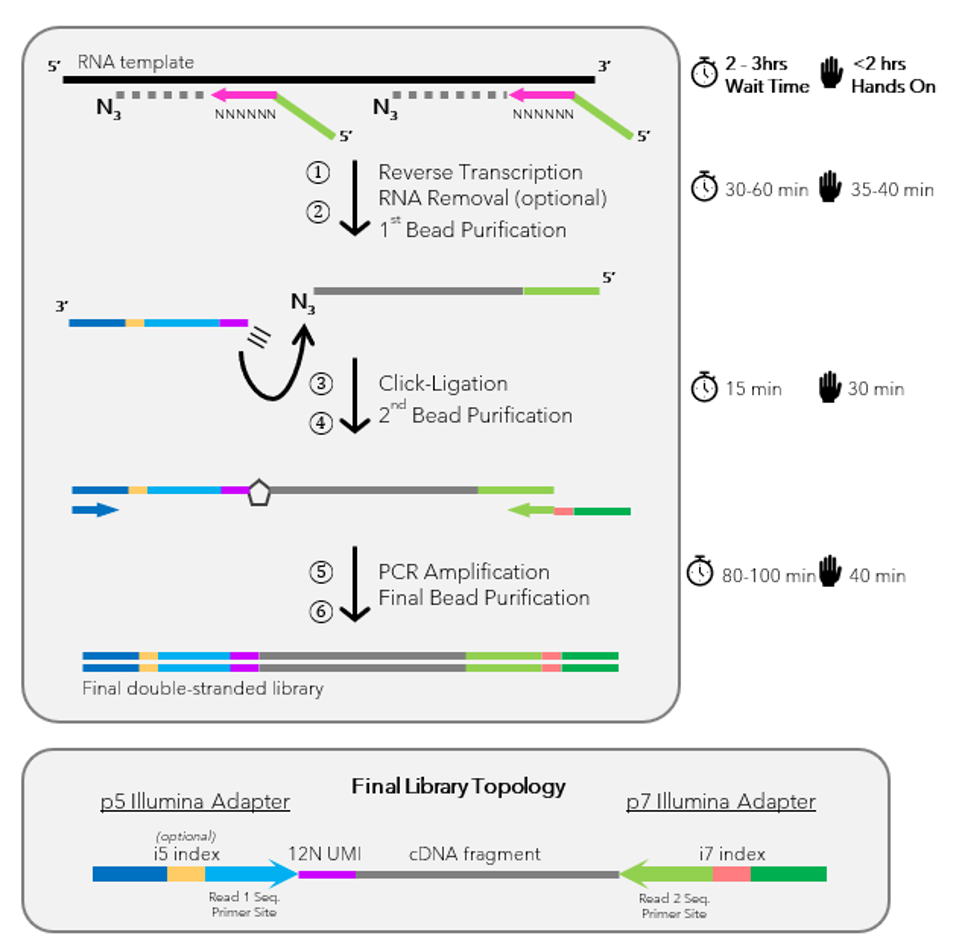
Kit Components
The RNA ClickSeq Library Preparation Kit includes:
- 3′-Azido-ddATP, -ddCTP, -ddGTP, -ddTTP
- Reverse transcription buffer and reagents
- Alkyne-modified sequencing adapters
- Click reaction buffer and Cu(I) catalyst
- PCR setup reagents (excluding primers)
Note: PCR primers are not included. A set of 12 compatible i7 index primers is available from baseclick. Custom primers can also be used.
Scientific background
ClickSeq builds on the principle of stochastic termination during reverse transcription, which results in a distribution of fragment lengths without the need for RNA fragmentation. The use of click chemistry for adapter ligation enhances specificity, reaction efficiency, and biocompatibility. The resulting triazole linkage is seamlessly read through by DNA polymerases during amplification.
This unique approach significantly reduces chimera formation and sequencing bias, as demonstrated by fewer than 3 aberrant events per million reads in comparative studies [1+2].
ClickSeq technology is also available for DNA sequencing, for sequencing of 3`polyadenylated RNA or for the usage with custom designed primers for reverse transcription reactions.
[1] Routh, A., Head, S. R., Ordoukhanian, P., & Johnson, J. E. (2015). ClickSeq: fragmentation-free next-generation sequencing via click ligation of adaptors to stochastically terminated 3′-azido cDNAs. Journal of molecular biology, 427(16), 2610-2616.
https://doi.org/10.1016/j.jmb.2015.06.011
[2] Jaworski, E., & Routh, A. (2017). ClickSeq: replacing fragmentation and enzymatic ligation with click-chemistry to prevent sequence chimeras. In Next Generation Sequencing: Methods and Protocols (pp. 71-85). New York, NY: Springer New York.
https://doi.org/10.1007/978-1-4939-7514-3_6
For a detailed list of publications utilizing ClickSeq technology please visit:
FAQ
-
What is ClickSeq?
ClickSeq is a platform and method for making Next-Generation Sequencing libraries. It is called ‘ClickSeq’ as it replaces the ligations and fragmentation steps common to NGS library preps with ‘Click-Chemistry’.
-
How does ClickSeq work?
ClickSeq follows the same principle of all Next Generation Sequencing (NGS) Library Prep Kits that seek to generate short fragments of cDNAs that are flanked with the appropriate sequencing adaptor for the user’s choice of sequencing platform (e.g. Illumina).
The process works by supplementing the Reverse Transcription step of an Next Generation Sequencing (NGS) Library Prep Kit with small amounts of terminating ‘azido-nucleotides’. These are stochastically incorporated into the cDNA during first stranded synthesis, yielding 3’azido-terminated cDNA molecules. The azido group is then ‘Click-Ligated’ to an alkyne-functionalized sequencing adaptor using Click-Chemistry (Copper Catalysed Azide-Alkyne Cycloaddition, CuAAC). The resultant click-linked single-stranded cDNA is then PCR amplified to generate a final sequencing-ready NGS library.
-
Can I sequence any RNA with ClickSeq?
Yes. Whatever you put into your NGS prep is what you will sequence, with the exception of very short RNAs. Since ClickSeq relies on the stochastic incorporation of azido-nucleotides into cDNA during reverse transcription, the reverse transcriptase might reach the end of small RNAs before an azido-nucleotide is incorporated. As a result, the cDNA cannot be click-ligated to the sequencing adaptor and thus will not be sequenced.
-
Can ClickSeq sequence DNA?
Yes. ClickSeq can sequence DNA with the exact same protocol as used for RNAseq provided that the RT enzyme has DNA-templated DNA polymerase activity (such as SuperScript III). Published examples of this can be found here:
https://academic.oup.com/gigascience/article/doi/10.1093/gigascience/giad009/7080818
-
Is ClickSeq published?
Yes. ClickSeq was originally published in 2015 by Routh et al in the Journal of Molecular Biology:
https://www.sciencedirect.com/science/article/abs/pii/S0022283615003514?via%3Dihub
ClickSeq has since been used in numerous publications, by the original inventors, by clients who have purchased ClickSeq kits and/or services as well as independent groups who have assembled their own reagents. See here for a curated list of publications:
-
Where can I find a protocol for ClickSeq?
Our protocol can be found here:
https://www.protocols.io/view/clickseq-random-primed-protocol-with-single-indexi-n92ld8jkov5b/v1
-
What do I need in addition to the kit to make a ClickSeq library?
Required third-party reagents include:
- SuperScript III™ Reverse Transcriptase, 200U/µL (Invitrogen; 18080-093 or 18080-044)
- ClickSeq Library Prep Kit Index Primers (baseclick, BCK-RNAseq-IP)
- [optional] RNaseOUT™ Recombinant Ribonuclease Inhibitor, 40U/µL (Invitrogen; 10777)
- OneTaq® 2X Master Mix with Standard Buffer (NEB; M0482S or M0482L) (Note: you must use OneTaq for this step, this enzyme cannot be substituted for a different PCR enzyme)
- [optional] RNase H, 5000 units/mL (NEB; M0297S or M0297L)
- SPRIselect (Beckman Coulter; B23317) or equivalent DNA/RNA Purification Beads (also known as SPRI beads)
- Nuclease free water
- 80% ethanol (made fresh)
-
Where can I find more information?
Please see an extensive set of FAQs:

