
Sensitive EdU Cell Proliferation Assay for Imaging
ClickTech Sensitive EdU Cell Proliferation Kit for Imaging
| Size | Catalog No. | Price |
|---|---|---|
| Dye 488 / 100 Assays | BCK-EdUPro488IM100 | € 525,00 |
| Dye 647 / 100 Assays | BCK-EdUPro647IM100 | € 525,00 |
Chemical Properties
-
Shelf Life
12 months unopened after receipt
-
Storage Conditions
2-8 °C
-
Physical State
kit system made of different components
-
CAS Number
n.a.
-
Excitation (max)
Dye 488: 496 nm | Dye 647: 643 nm
-
Emission (max)
Dye 488: 516 nm | Dye 647: 662 nm
-
Ɛ (max)
Dye 488: 83.000 cm-1M-1 | Dye 647: 250.000 cm-1M-1
-
Preparation/Handling
please see user manual of the kit
Product Information
High-sensitivity detection of DNA synthesis for fluorescence microscopy
The ClickTech Sensitive EdU Cell Proliferation Kit for Imaging enables precise visualization of DNA replication in cultured cells using EdU (5-ethynyl-2′-deoxyuridine) incorporation and copper-catalyzed click chemistry. This assay is optimized for fluorescence microscopy and provides high signal intensity with minimal background, making it suitable for cell cycle studies, proliferation analysis, and multiplexed imaging.
EdU is incorporated into DNA during the S-phase and detected via a bioorthogonal reaction with picolyl-azide fluorophores. The picolyl group enhances reaction kinetics and signal strength while allowing for reduced copper concentrations, which helps maintain cell morphology and viability.
Fluorescent Dye Options
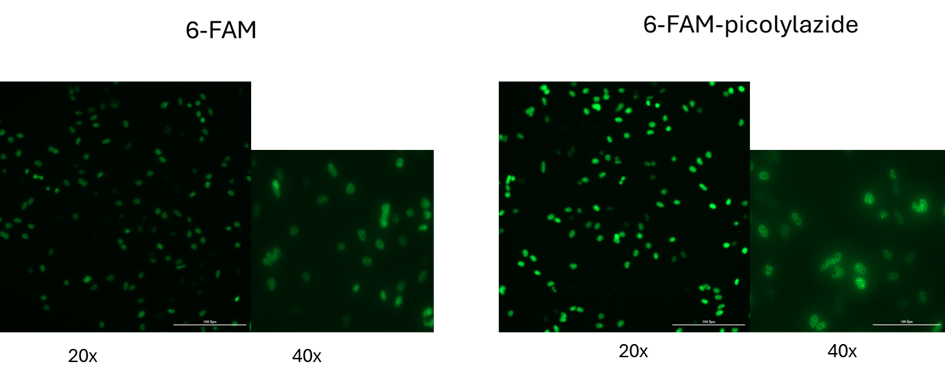
- 6-FAM-Picolyl-Azide (Green Channel): Suitable for co-staining with DAPI and FITC-compatible filter sets
- Eterneon-Red 645-Picolyl-Azide (Far-Red Channel): Ideal for deep-tissue imaging and samples with high background fluorescence
Why Choose the Sensitive EdU Assay?
Direct Detection of DNA Synthesis: EdU incorporation provides a direct readout of DNA replication without the need for DNA denaturation or antibody-based detection.
High Signal-to-Noise Ratio: Picolyl-azide fluorophores yield strong, stable fluorescence with low background, suitable for imaging low-proliferation samples. (Up to 40× stronger signal intensity)
Low Cytotoxicity: The click reaction is formulated with reduced copper levels to minimize cellular stress and preserve sample integrity.
Fast Protocol: The assay can be completed in under 2 hours, including EdU labeling, fixation, permeabilization, and detection.
Multiplexing Compatibility: Compatible with nuclear stains (e.g., DAPI), viability markers, and antibody-based phenotypic markers for multicolor imaging.
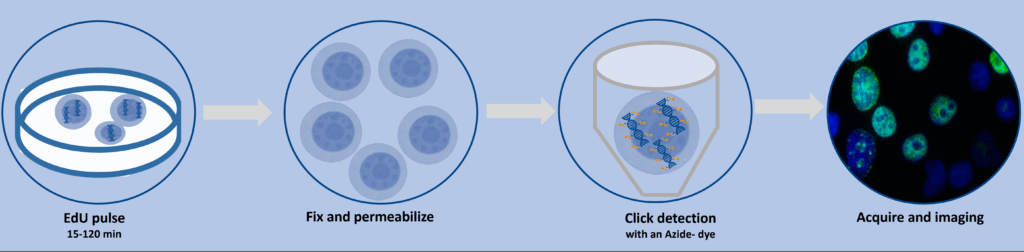
Workflow Summary
- EdU Labeling
Incubate cells with EdU (typically 10 µM) for 15–120 min depending on cell type and experimental goals. - Fixation & Permeabilization
Use formaldehyde-based fixation and mild detergents to preserve morphology and allow dye access. - Click Reaction
Add the picolyl-azide dye cocktail and incubate under gentle conditions. Wash thoroughly. - Optional Co-Staining
Add nuclear stains or antibody-based markers for phenotypic analysis. - Microscopy Readout
Visualize EdU-positive nuclei using appropriate filter sets. Quantify signal intensity or cell counts using image analysis software.
LITERATURE
Divergent Synthesis of Ultrabright and Dendritic Xanthenes for Enhanced Click-Chemistry-Based Bioimaging. 2023, Chemistry – A European Journal.
https://doi.org/10.1002/chem.202202633
A Genetically Encoded Picolyl Azide for Improved Live Cell Copper Click Labeling.” Front. Chem., 2021.
https://doi.org/10.3389/fchem.2021.768535
Efficient Tandem Copper‐Catalyzed Click Synthesis of Multisugar‐Modified Oligonucleotides. Angewandte Chemie International Edition, e202405161.
https://doi.org/10.1002/anie.202405161
FAQ
-
How does the picolyl-azide modification improve imaging performance compared to standard azides?
The picolyl group enhances the copper-catalyzed click reaction by increasing reaction kinetics and efficiency. This results in significantly stronger fluorescence signals (up to 40×), reduced background, and improved labeling consistency—especially important for low-proliferation samples or high-resolution imaging.
-
Can this assay be used for quantifying proliferation in 3D cultures or tissue sections?
Yes. The far-red dye (Eterneon-Red 645-Picolyl-Azide) is particularly well-suited for thick samples and 3D cultures due to its low autofluorescence and deep tissue penetration. Ensure proper fixation and permeabilization protocols are optimized for your sample type.
-
What is the recommended copper concentration for the click reaction, and how does it affect cell morphology?
The assay uses a reduced copper formulation (typically <100 µM CuSO₄), which minimizes cytotoxicity and preserves cellular and nuclear morphology. This is especially beneficial for sensitive cell types and downstream imaging applications.
-
Is the assay compatible with high-content screening (HCS) platforms?
Yes. The assay is fully compatible with HCS systems using standard FITC or Cy5 filter sets. The dyes are photostable and produce consistent signal intensities across wells, making them suitable for automated image acquisition and quantitative analysis.
-
How should EdU pulse duration be optimized for different cell types?
Pulse duration depends on the cell cycle dynamics of the model system. For rapidly dividing cells, a 30–60 min pulse may be sufficient. For slow-cycling or stem cells, longer pulses (up to 2 h) may be needed to capture cumulative DNA synthesis. Always validate pulse conditions experimentally.
-
Can this assay be combined with antibody-based immunofluorescence?
Yes. The mild fixation and permeabilization conditions preserve antigenicity, allowing co-staining with antibodies targeting cell cycle markers (e.g., Ki-67, phospho-H3), differentiation markers, or apoptosis indicators.
-
What are the best practices for minimizing background fluorescence in imaging?
- Use freshly prepared dye cocktails and avoid prolonged incubation.
- Include dye-only controls to assess non-specific binding.
- Use appropriate blocking steps if combining with antibody staining.
- Optimize washing steps to remove unbound dye and reduce residual copper.

