Poly RNA is a homopolymeric RNA, meaning it is a strand made entirely of repeating units of the same nucleotide base. Examples include poly(C), poly(G),poly(U) and poly(A). Unlike natural messenger RNAs, which have a varied sequence of bases, poly RNAs are usually synthetic constructs created in laboratories using specialized RNA polymerases or chemical synthesis.
Long poly RNAs are uncommon in living cells, though short homopolymer tracts can occur naturally and influence transcription, RNA stability, and molecular interactions.
Despite their simple composition, poly RNAs have played a significant role in molecular biology, from assisting in the decoding of the genetic code to advancing modern RNA-based technologies. Their uniform sequence makes them ideal for studying the impact of RNA composition on transcriptional activities, structural stability and downstream biological or technological applications.
Role in Molecular Biology
- Transcription Context: Although cells generally do not transcribe long homopolymeric RNAs, short sequences of identical nucleotides can be found in natural transcripts. These sequences can influence transcriptional regulation by affecting the movement or pausing of RNA polymerase.
- RNA Stability: Homopolymer stretches can alter the secondary structure of RNA and its affinity for binding proteins, which can affect its stability and degradation rates. For instance, certain repeats may encourage protective folding, or conversely, render the RNA more vulnerable to nucleases.
- Downstream Applications:
- Genetic Code Decoding: Historically, synthetic poly RNAs have been crucial in establishing the correspondence between codons and amino acids in cell-free translation systems.
- Biophysical & Structural Studies: They serve as model systems for understanding RNA folding, base pairing and RNA–protein interactions.
- Nanotechnology & Therapeutics: They have been explored for their ability to form stable, programmable structures for use in drug delivery, biosensing and synthetic biology.
What is Poly RNA?
Poly RNA refers to homopolymeric RNA molecules strands consisting entirely of a single type of nucleotide, such as poly-A, poly-U, poly-C or poly-G.
Poly RNA can occur naturally, for instance as poly(A) tails in eukaryotic mRNA or U-rich termination signals in certain transcripts. It can also be produced synthetically for research purposes.
Their simplicity makes them valuable tools for studying the effects of RNA sequence on transcription, stability, and molecular interactions. Poly-A and poly-U in particular have well-established roles in post-transcriptional regulation.
Poly RNA sequences can be either naturally occurring or synthetically produced.
Natural poly RNA:
- Poly(A) Tails in mRNA: messenger RNA (mRNA) molecules in eukaryotic cells typically have a polyadenylated tail(poly(A) tail) at their 3′ end which is added post-transcriptionally.
- Viral Genomes: some viruses produce RNAs with poly(A) or poly(U) sequences to mimic host mRNA or regulate replication.
- Non-coding RNAs: certain non-coding RNAs may contain homopolymeric stretches that affect their structure and the way they interact with other molecules.
Researchers can produce synthetic poly RNA in the lab:
- Chemical Synthesis: short poly RNA sequences can be synthesized using solid-phase synthesis techniques.
- In Vitro Transcription: longer poly RNA molecules can be generated using RNA polymerases (e.g., T7, SP6) and DNA templates with homopolymeric regions.
- Enzymatic Methods: Poly(A) polymerase can be used to add poly(A) tails to RNA molecules in vitro post-transcriptionally.
Biological Role of Poly‑A and Poly‑U
- Poly-A sequences: These form the poly(A) tail at the 3′ end of eukaryotic mRNA, which is essential for the stability of the mRNA, its export from the nucleus, and efficient translation, through interactions with poly(A)-binding proteins.
- Poly-U sequences: These often act as intrinsic transcription terminators in prokaryotes and participate in RNA decay pathways. In these pathways, U-rich regions help to recruit the machinery responsible for degradation.
Poly-U RNA and Its Research Significance
What is Poly-U RNA?
Poly-U RNA refers to uridine-rich RNA sequences consisting of an uninterrupted series of uridine nucleotides (U). These homopolymeric RNAs can occur naturally, for instance as U-rich tracts in certain bacterial transcripts or mitochondrial RNA processing signals, or as part of intrinsic transcription terminators. Alternatively, they can be produced synthetically in a laboratory setting using RNA polymerases with homopolymeric templates or via chemical synthesis.
Whether natural or synthetic, poly-U RNA serves as both a biological signal and a powerful experimental tool, enabling discoveries in RNA–protein recognition, transcript stability and the fundamental rules of genetic coding:
- Studying RNA–Protein Interactions:
- U-rich motifs are recognised by specific RNA-binding proteins (RBPs), including HuR, TTP and others, which are involved in post-transcriptional regulation.
- poly-U RNAs are used in experiments to investigate how RBPs identify sequence motifs and influence splicing, mRNA stability and translation efficiency.
- RNA Decay Mechanism:
- In bacteria: Poly-U tracts function as part of intrinsic transcription terminators, destabilising RNA–DNA hybrids and promoting transcript release.
- In eukaryotes: U-rich elements, which are often found in the 3′ untranslated region (UTR), recruit RNA degradation machinery. This links poly-U sequences with mRNA turnover and quality control pathways
- Experimental RNA Analogs:
- Historically, synthetic poly-U RNA played a key role in deciphering the genetic code by demonstrating that UUU encodes phenylalanine.
- Today, poly‑U remains a versatile model for:
- Studying RNA secondary structure formation.
- Testing hybridization with complementary RNAs (e.g., poly‑A).
- Exploring interactions with enzymes such as polymerases, nucleases, and RNA‑modifying proteins.
Applications of Poly-U RNA
Poly-U RNA has played a pivotal role in molecular biology. It has served as both a historical breakthrough tool and a modern experimental model for exploring translation, ribosome function and RNA turnover.
- Ribosome‑Binding Studies: Synthetic poly-U RNA was an early tool used to study how ribosomes bind to RNA. Its repetitive sequence provides a clear model for measuring ribosome activity without any structural interference.
- In Vitro Translation Assays: In early cell-free experiments, poly-U RNA caused ribosomes to produce chains consisting only of phenylalanine, revealing that the codon for phenylalanine is UUU. This key discovery helped to decode the genetic code, and the technique is still used to test translation today.
- RNA Stability and Degradation: U-rich sequences act as natural signals for RNA turnover. They can either recruit decay machinery or influence exonuclease accessibility. This makes poly-U RNA a valuable model for studying the effect of sequence composition on RNA half-life, degradation pathways and quality control mechanisms in both prokaryotic and eukaryotic systems.
Poly-A RNA Isolation Techniques
Poly-A RNA Isolation Techniques
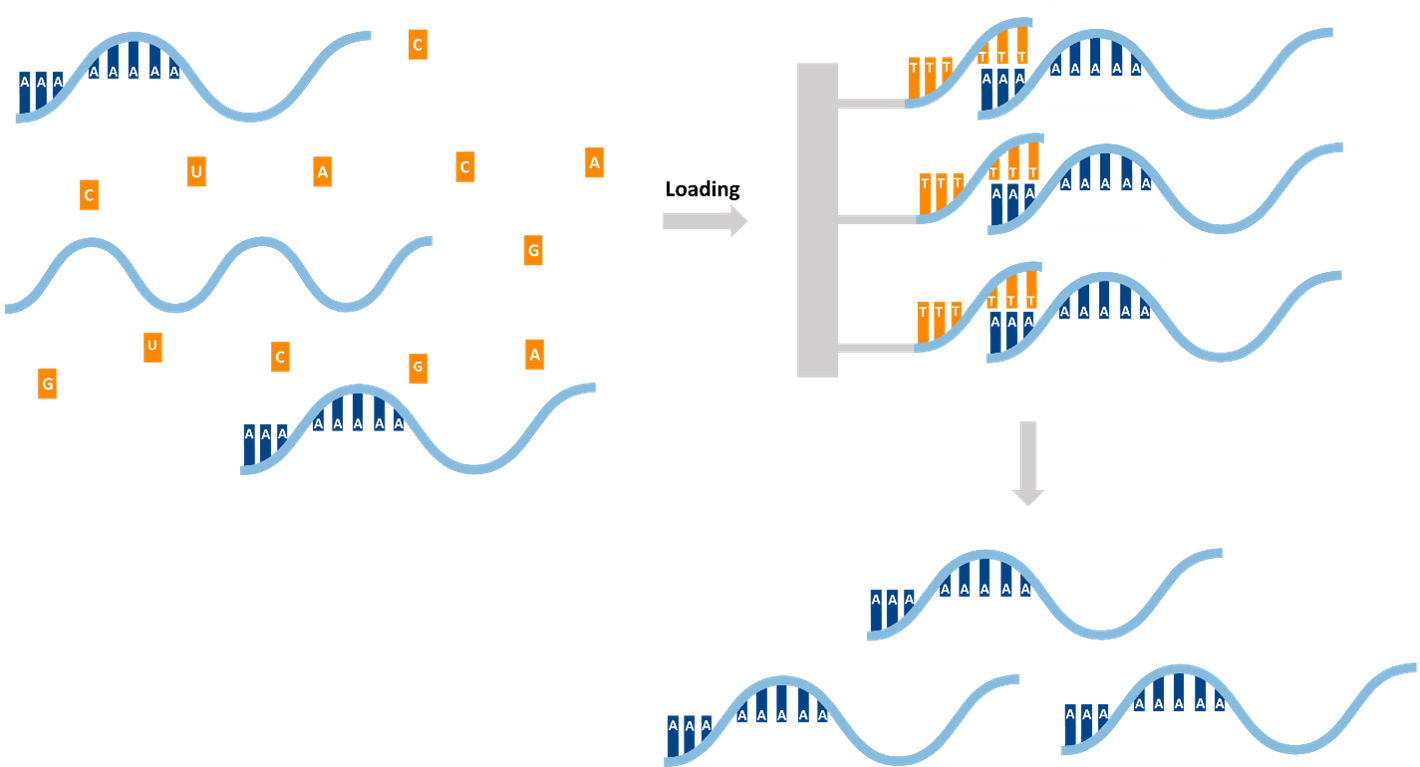
Researchers often isolate poly-A RNA because the polyadenylated tail at the 3′ end is characteristic of mature eukaryotic mRNAs. This tail is added during post-transcriptional processing and is largely absent from other abundant RNA species, such as ribosomal RNA (rRNA) and transfer RNA (tRNA). Using oligo(dT) probes or beads that specifically bind to poly(A) tails enables scientists to selectively enrich mRNA from total RNA, which otherwise contains a high percentage of rRNA and other non-coding RNAs.
This enrichment yields a population that predominantly comprises protein-coding transcripts, enabling researchers to focus on the abundance, diversity and processing of mRNA rather than the entire RNA pool. This selective capturing improves sequencing efficiency and reduces background noise. It is also essential for applications such as transcriptomics, RNA sequencing (RNA-Seq) and gene expression studies, where the aim is to obtain an accurate and clear view of the actively expressed, fully processed transcriptome.
Isolation Methods
The isolation of polyadenylated mRNA is a crucial step in transcriptomics workflows, as it enables the enrichment of protein-coding transcripts while removing most ribosomal and non-polyadenylated RNAs. Two widely used approaches exploit oligo(dT)-poly(A) hybridization: magnetic bead-based captureing and column based purification.
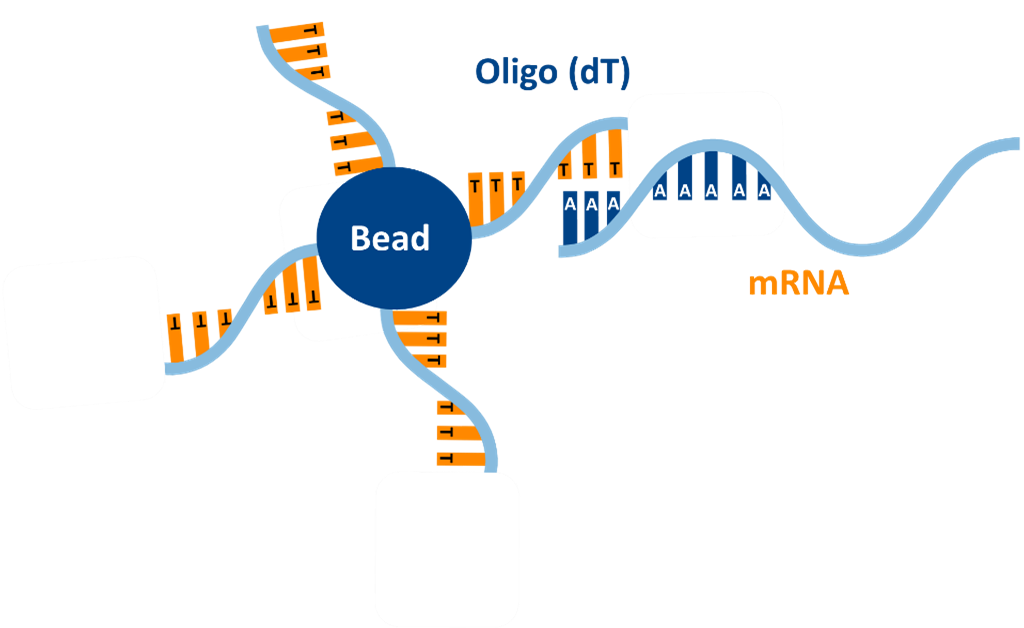
- Oligo‑dT Magnetic Beads: Oligo-dT beads bind to the poly(A) tails of mature mRNA, enabling the selective capture of mRNA from total RNA pools. Following magnetic separation and washing, the purified mRNA can be eluted with high specificity and integrity. This scalable, automation-friendly method removes over 99% of rRNA, although yield depends on the quality of the input and excludes small or non-polyadenylated RNAs.
- Column Purification with Oligo‑dT Matrices: In column purification with oligo-dT matrices, silica supports are used to bind poly(A) mRNA and wash away other RNAs. This method can handle large volumes of RNA efficiently and often yields more mRNA than bead methods. Although the purity is high, incomplete binding may result in the presence of rRNA, so gentle handling is required to protect the integrity of the RNA.
baseclick’s Role in Poly RNA Research
baseclick GmbH offers click-chemistry-based tools for the efficient research of poly(A) RNA, from the profiling of natural mRNA to the generation of synthetic transcripts. The Poly(A)-ClickSeq Library Prep Kit enables 3′ end-focused RNA-Sequencing of polyadenylated RNA using oligo-dT primers for reverse transcription and azido-terminated nucleotides to precisely map polyadenylation sites, study alternative polyadenylation (APA) and analyse 3′ untranslated region (UTR) usage without fragmentation or ribodepletion working with as little as ~10 ng of RNA, even if degraded. The ClickSeq Library Prep Kit uses the same fragmentation-free, ligation-free chemistry for sequencing RNA or DNA. It supports oligo-dT priming for mRNA-Seq and minimises artefacts for applications such as gene expression and splice isoform analysis. For the production of synthetic RNA, the IVT High Performance T7 RNA Synthesis Kit produces high-purity, full-length mRNA with the option of co-transcriptional capping and the use of modified nucleotides. This makes it ideal for the generation of poly-A RNA controls or analogues. The ClickTech Oligo Link Kit enables precise CuAAC ligation of oligonucleotides or functional tags and is useful for creating custom oligo-dT constructs or primers. Together, these platforms provide integrated, highly specific solutions for isolating, profiling and engineering poly-A RNA.

