2′,3′-dideoxythymidine triphosphate, ddTTP, is a synthetic nucleotide analog used in molecular biology as a chain-elongating inhibitor of DNA Polymerase. It is used in the Sanger method for DNA sequencing. It lacks hydroxyl groups (OH groups) at both the 2′ and 3′ positions of the sugar moiety compared to ribose, enabling to prevent the formation of new phosphodiester bonds, thereby terminating DNA strand elongation if incorporated during DNA synthesis. This nucleotide is commonly labeled or used alongside other deoxyribonucleotides (dNTPs) to generate DNA fragments of different lengths for analysis.
What is ddTTP?
ddTTP is similar to the natural nucleotide dTTP (deoxythymidine triphosphate) but has a critical structural difference.
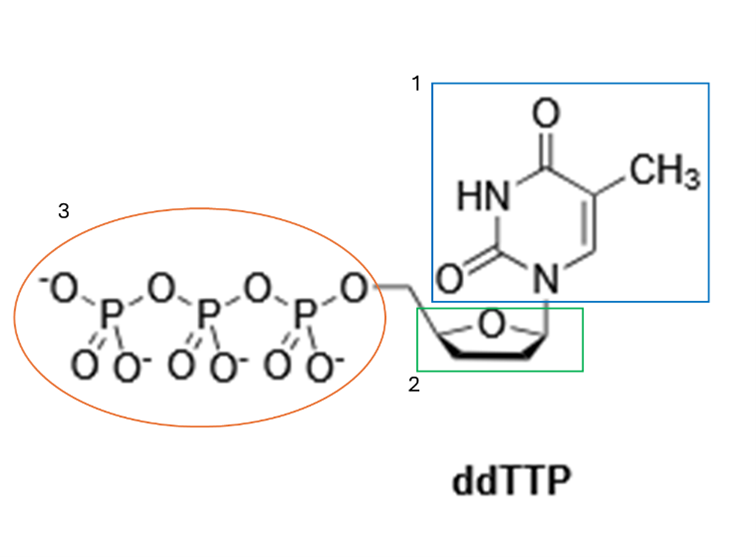
Chemical Structure
- Base: Thymine (1)
- Sugar: Modified deoxyribose that lacks both the 2′-OH and the 3′-OH groups (2)
- Triphosphate group (3)
Key Differences Between ddTTP and dTTP
- dTTP has a 3′-OH group on the sugar, which is essential for forming the phosphodiester bond to the next nucleotide during DNA elongation.
- ddTTP lacks the 3′-OH group. Once it is incorporated into a growing DNA strand, it causes chain termination, meaning that no further nucleotide can be added.
The role of ddTTP in DNA sequencing
Sanger sequencing, also known as the chain termination method, is a DNA sequencing technique developed by Frederick Sanger in 1977.
Basic Principle
Sanger sequencing is a technique used to determine the nucleotide sequence of DNA. It involves synthesizing complementary strands in the presence of normal dNTPs and small amounts of ddNTPs, such as ddTTP. ddNTPs lack the 3′–OH group essential for forming phosphodiester bonds with the next nucleotide.
How ddTTP Causes Termination
- Normal DNA synthesis uses dNTPs, which allow DNA elongation due to the 3′-OH group in the sugar ring.
- When a ddTTP is incorporated instead of a normal dTTP, the DNA polymerase cannot add any more nucleotides due to the lack of a 3′–OH group. This leads to the termination of DNA strand elongation at that point.
Generation of DNA Fragments
- As the sequencing reaction includes a mixture of all four dNTPs and a small amount of one ddNTP (e.g., ddTTP),the DNA polymerase randomly incorporates the ddTTP at different positions where thymine would normally be added.
- This results in the accumulation of DNA fragments of various lengths, each of which ends at a position where a ddTTP nucleotide was incorporated.
Sequence Analysis
- The fragments can be separated by size using gel or capillary electrophoresis.
- In modern automated systems each ddNTP is labeled with a distinct fluorescent dye.
- The DNA sequence can be reconstructed by identifying the color and length of each fragment.
baseclick offers azide-modified 3′-Azido-2′,3′-ddTTP for precision termination for Sanger Sequencing, meanwhile allowing for modification of the azido group by Click Chemistry.
How It Works in Sanger Sequencing
- Like standard ddTTP, 3′-Azido-ddTTP lacks the 3′-OH group.
- When 3′-Azido- ddTTP is incorporated by DNA polymerases, it terminates DNA synthesis. This makes it suitable for chain-termination sequencing.
- The azido group at the 3′ position can be used to attach fluorescent dyes, biotin or other tags via click chemistry (e.g., CuAAC or SPAAC).
Learn more about Click Chemistry here: Click Chemistry Glossary
The chain termination mechanism
The absence of a 3′-OH group in ddTTP prevents DNA polymerase from forming a phosphodiester bond with the next incoming nucleotide.
Why Polymerase Stops
- DNA polymerase adds nucleotides to the 3′-OH group of the growing DNA strand. The 3′-OH on the terminal nucleotide of the growing DNA chain acts as a nucleophile to attack the 5′-triphosphate of the incoming dNTP, linking them together and extending the chain.
- ddTTP lacks 3′-OH and has only a hydrogen at the 3′ position. The DNA chain cannot be extended further, stopping polymerase activity at that point.
- Once ddTTP is incorporated, no further nucleotide can be added, effectively terminating the DNA strand.
Principle of Controlled Termination
In Sanger sequencing, ddNTPs (such as ddTTP) are incorporated alongside regular dNTPs in a reaction mixture to control the termination of the DNA strand. Termination occurs randomly but specifically when a ddNTP is added instead of a dNTP, producing DNA fragments that end at every position that corresponds to that base. The dNTP:ddNTP ratio is adjusted to control this process and ensure that fragments of varying lengths are generated without the reaction being exhausted prematurely.
This creates a precise “sequence ladder” of fragments, each ending at a specific base. The DNA fragments can be separated by size via electrophoresis, which allows for reliable determination of the nucleotide order and mapping of the sequence.
Applications of ddTTP in molecular biology
Classical Sanger sequencing, also known as chain-termination sequencing, is a fundamental method for determining the nucleotide sequence of DNA fragments. It uses ddNTPs, including ddTTP, which lack a 3′-OH group to terminate the DNA strand at a specific base.
Sanger sequencing is used to analyse genes and mutations in order to precisely identify genetic variations, such as single-nucleotide polymorphisms (SNPs), insertions, deletions or point mutations. This is achieved by comparing the sequenced DNA to reference genomes.
In modern automated sequencing systems detection is enhanced by the fluorescent labeling of ddNTPs, including ddTTP. Each ddNTP (A, T, G, C) carries a distinct fluorophore, enabling automated detection via capillary electrophoresis. As the labeled fragments migrate through a capillary gel and pass a laser detector, the emitted fluorescence identifies the terminating base at each position. This enables high-throughput, real-time sequence readout on computerized systems.
Sanger sequencing is crucial in diagnostics for confirming disease-causing mutations. For example, it is used to assess cancer risk by examining BRCA genes. This technique is the gold standard in research for validating next-generation sequencing (NGS) results and for studying small genomic regions. In personalized medicine, Sanger sequencing guides targeted therapies, such as the selection of drugs based on patient-specific genetic profiles in oncology and pharmacogenomics. Although it has largely been replaced by NGS for large-scale applications, its accuracy and simplicity make it indispensable for clinical validation and low-throughput needs.
baseclick offers ClickSeq DNA Library Prep Kit that utilizes the unique chain-terminating properties of 3′-azido-2′,3′-dideoxynucleotides, including 3′-Azido-2′,3′-ddTTP, for efficient, accurate, and click-enabled DNA library preparation for Illumina Next Generation Sequencing. There are also variants available for RNA sequencing or for sequencing the Poly A region of mRNA.
- Fast & reliable: Simplified protocol for high-quality libraries
- Click chemistry ready: Enables post-ligation labeling and multiplexing
- Ideal for NGS & targeted sequencing
- Perfect for mutation analysis, diagnostics, and personalized medicine
DNA-ClickSeq – where smart chemistry meets powerful sequencing.
How ddTTP is used practically in the laboratory
Practical Use of ddTTP in Sanger Sequencing
- The typical reaction mixture (10 – 20 µl) includes:
| Component | Purpose | Concentration |
| Template DNA | Target | 10–100 ng |
| Primer | Binding | 5–10 pmol |
| Polymerase | Extension | 0.5–1 U |
| dNTPs | Building blocks | 200 μM ea |
| Fluorescent ddNTPs | Terminators; ddTTP | 1–8 μM ea. (~1:100 ratio) |
| Buffer | Enzyme support | 1× |
- PCR Setup with dNTPs and ddTTPs:
- The reaction is similar to PCR, designed for linear amplification.
- Fluorescent labeled ddTTP is added in low amounts relative to dTTP.
- DNA polymerase randomly incorporates ddTTP at thymine positions, which leads to chain termination.
- This generates a mixture of DNA fragments, each ending at a different thymine.
- Visualization and Data Interpretation
- Fragments are separated by gel or capillary electrophoresis.
- The fluorescent labeled ddTTP -terminated fragment emits a signal.
- A sequencer detects fragment lengths and fluorescence, which enables the reconstruction of the DNA sequence.
- The result is displayed as a chromatogram showing the peaks for each base in the sequence.
- It reveals mutations by comparing with a reference, with an accuracy of >800 bp/read and 99.9%.
Total time: 2–4 hrs for 96 samples.
Differentiating ddTTP from other nucleotide analogues
All ddNTPs lack 3′-OH for chain termination, but differ by base:
| ddNTP | Terminates at | Detects base |
| ddATP | Adenine (A) | T in template |
| ddCTP | Cytosine (C) | G in template |
| ddGTP | Guanine (G) | C in template |
| ddTTP | Thymine (T) | A in template |
Key Difference: ddTTP uniquely terminates at A sites, labeling T positions.
Unique Role in Sequencing Thymine Bases:
- Incorporation: DNA polymerase adds ddTTP opposite A in template → stops extension.
- Ladder Generation: Low [ddTTP] creates fragments ending in T at every A position.
- This allows precise identification of A bases in the template strand.
Impact on Sequencing Accuracy
- Each ddNTP is incorporated selectively and efficiently to ensure accurate base calling.
- The balance between dNTPs and ddNTPs is a critical issue. Too much ddTTP can cause premature termination, while too little may miss termination points.
- Proper use of ddTTP contributes to high-resolution sequencing, especially in regions rich in adenine.
Using equal ddNTP ratios ensures uniform termination and a high-quality electropherogram. Omitting ddTTP fails to terminate ~25% of bases.
baseclick offers high-quality azide-modified ddNTPs that closely resemble natural nucleotides. This enables chemoenzymatic labelling of single-stranded DNA (ssDNA) for successful sequencing assays:
Why ddTTP remains essential in DNA sequencing
ddTTP: A Key to Precision in Genetic Analysis
ddTTP plays a critical role in Sanger sequencing, enabling the accurate identification of thymine bases. This allows the termination of DNA strand elongation at specific points and the synthesis of opposite adenine. Selective incorporation ensures the generation of DNA fragments that reflect the exact sequence of the template.
In research and diagnostics, ddTTP contributes to:
- Reliable mutation detection
- High-quality sequencing results
- Accurate genotyping and gene analysis
Its use in fluorescent labeling further enhances detection in automated systems, making ddTTP essential for personalized medicine, clinical diagnostics, and genetic research.



