FACS cell sorting: targeted isolation of cells using fluorescence markers
Fluorescence Activated Cell Sorting (FACS) is an advanced, laser-based technique used to separate and analyze specific cell types from complex biological mixtures such as blood, tissue, or cultured cell suspensions. By using fluorescently labeled antibodies that bind to distinct molecular markers, FACS enables the isolation and collection of highly defined, viable cell subpopulations with exceptional precision and purity.
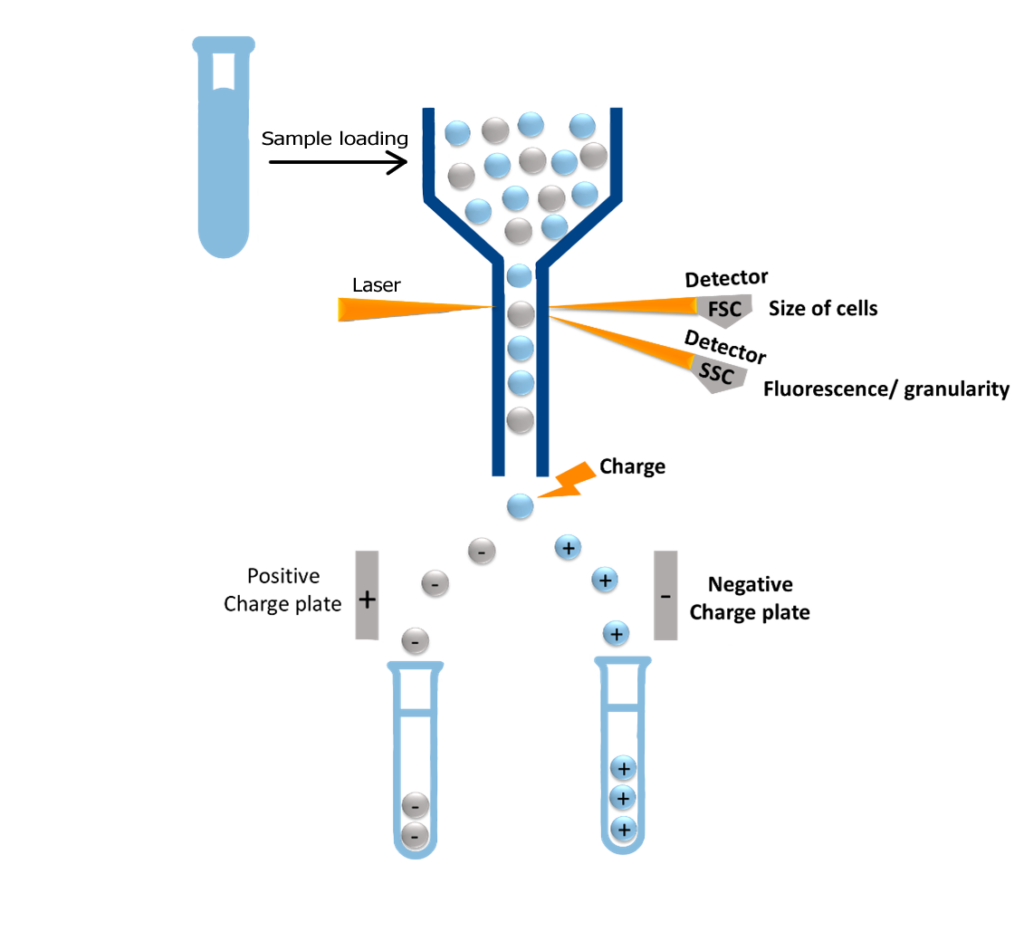
FACS Cell Sorting is a specialized form of flow cytometry, integrating fluorescence detection with physical sorting. Here’s how the process works:
– Fluorescent Tagging: Cells are stained with antibodies conjugated to fluorochromes that target specific molecular features.
– Single-Cell Stream: In a liquid flow, these labeled cells pass one-by-one through a focused laser beam inside the flow cytometer.
– Signal Detection: The laser excites the fluorochromes, and detectors measure light scatter and fluorescence intensity cell by cell.
– Electrostatic Sorting: Droplets containing a single cell are assigned an electrical charge. These single-cell-containing charged droplets, when the cell meets defined fluorescence criteria, are deflected by an electrostatic field into designated collection containers.
FACS Cell sorting is a cornerstone in contemporary biological and medical science due to its unparalleled accuracy and versatility. It is commonly used for:
- Immunology: Immunophenotyping and isolation of immune cell subsets
- Stem Cell and Regenerative Medicine: Purification of stem cells or engineered cells for therapeutic use
- Cancer Research: Analysis of tumor heterogeneity and rare cancer cell populations, for example in the blood
- Cell Biology: Investigation of cell cycle, apoptosis, and gene expression at single-cell resolution
- Drug Discovery: High-throughput screening and functional studies of cell responses
FACS Cell Sorting, empowers researchers to dissect complex biological systems with high sensitivity, specificity, and reproducibility, enabling profound insights into cellular processes, disease mechanisms, and therapeutic potential.
Where FACS cell sorting is applied: beyond immunology
FACS Cell Sorting is essential across biological research, enabling precise cell sorting and analysis.
Immunology
Fluorescence Activated Cell Sorting is a key tool for identifying and isolating specific immune cell types, such as T cells, B cells, natural killer (NK) cells or dendritic cells. These cell types can be precisely filtered and separated from heterogeneous samples using fluorescence-labeled antibodies that selectively bind to their surface markers. Therefore, FACS is a valuable technique for immunological research and diagnostics.
Cancer Research
FACS Cell Sorting is used to detect and specifically enrich tumor cells based on surface or intracellular markers. This technique allows for a thorough examination of cancer cell populations in blood, tissue, or cell cultures, even when they are present in small quantities. DNA dyes, such as propidium iodide, and apoptosis markers can be used to determine cell cycle status and cell viability in order to evaluate responses to cancer treatment or resistance mechanisms by FACS.
Stem Cell Studies
FACS allows for the targeted isolation of stem cells from complex cell mixtures, such as blood or tissue samples. Characteristic surface markers are used to select either pluripotent or differentiated cell types. This allows for the isolation of pure stem cell fractions for research and therapeutic applications.
Microbiology
Fluorescence Activated Cell Sorting has also gained importance in microbiology. Microorganisms are labeled with either nonspecific DNA dyes (e.g. SYBR Green) or specific antibodies to sort them according to their fluorescence profiles. This allows, for example, the targeted enrichment of pathogenic bacteria or the investigation of the physiological states of microbial subpopulations. FACS also supports the analysis of bacterial biofilms and of virus-infected cell populations.
FACS Cell Sorting stands out for its ability to deliver exceptionally pure cell populations, often achieving 95–99% purity from even highly complex mixtures, which makes its precision not only impressive but transformative. By utilizing multi-parameter tagging and advanced gating, FACS pinpoints specific cells based on surface markers, size, and granularity. It also ensures recovery of live, viable cells that are fully functional for sensitive downstream assays or clinical applications. This high-purity sorting minimizes background noise, enhancing signal clarity and boosting reproducibility in subsequent experiments.
How FACS cell sorting works: basic principles
Detection & labeling
At the heart of FACS Cell Sorting is the use of fluorescently labeled antibodies or dyes that bind to specific cell surface markers, typically proteins like CD antigens that define distinct cell types or functional states. Researchers conjugate these antibodies with fluorescent dyes such as FITC, PE, or APC to selectively label targeted cells.
As each cell flows individually through a laser beam in the FACS instrument, the attached fluorochromes emit light at characteristic wavelengths. These emissions are captured and quantified, enabling the system to identify, analyze, and sort cells with remarkable precision. This labeling strategy supports multiparametric sorting, allowing simultaneous detection of several markers and making it possible to distinguish even closely related or rare cell populations.
Ultimately, it’s this fluorescence-based discrimination that empowers FACS to deliver exceptionally pure, viable cell subsets tailored for downstream applications from gene expression profiling to immunotherapy and regenerative medicine.
Detection & labeling
FACS Cell Sorting is a sophisticated technique that combines flow cytometry with precision sorting to isolate live, pure cells from complex samples through following process:
- Fluidics & Single-Cell Flow
The fluidic system ensures that cells flow individually and uniformly through the measuring chamber: cells are suspended in fluid and guided through a narrow nozzle via hydrodynamic focusing, aligning them into a single-file stream so they pass individually through the laser interrogation point.
- Laser-Based Detection
As each cell enters the path of one or more lasers:
– Fluorescent antibodies or dyes bound to cell surface markers (e.g., CD antigens) are excited and emit light.
– Light scatter is measured, forward scatter reveals cell size, while side scatter indicates granularity.
– Fluorescence intensity & wavelength are detected by sensors, revealing marker expression and cellular identity.
- Real-Time Classification
The electronics system processes these signals in real time and controls cell sorting. The system compares each cell’s signal to gating parameters (e.g., CD4⁺CD25⁺) to determine whether it matches the target population.
- Electric Charge Sorting
Each analyzed cell is enclosed in a tiny fluid droplet:
– If the cell meets sorting criteria, the droplet receives a positive or negative electric charge.
– Droplets pass through electrostatic deflection plates, which steer them into designated collection tubes.
– Uncharged droplets, representing unwanted cells, flow straight into the waste container.
Benefits of FACS cell sorting in research workflows
FACS Cell Sorting offers an unmatched combination of high purity, viability, and direct usability, making it a linchpin of both research and clinical science.
FACS routinely delivers >95–99% purity, even from heterogeneous samples, by applying multiparametric gating with fluorescent antibodies and dyes that bind specifically to surface markers. A key advantage of fluorescence-activated cell sorting is the ability to analyze numerous parameters per cell simultaneously, such as surface proteins, intracellular signals, DNA content, and metabolic activity. Depending on the instrument configuration, 10 or more fluorescence channels can be analyzed at once, providing a detailed cellular profile. This precision ensures minimal contamination and is especially crucial when isolating rare cell populations for downstream assays.
Moreover, the gentle nature of FACS sorting cells within fluid droplets without physical damage, preserves cell viability and functional integrity. That’s a major win for applications needing live cells, such as culture expansion, proliferation tracking, and therapeutic use.
Sorted cells are immediately compatible with a wide spectrum of downstream workflows, including gene expression analysis (qPCR, RNA-seq), proteomics, single-cell sequencing, cytokine release assays, and even in vivo transplantation. FACS merges specificity with functionality, allowing scientists to confidently move from complex biological mixtures to pure, living populations tailored for discovery or therapy.
FACS Cell Sorting, excels as a high-performance tool in cell biology by combining exceptional purity, multiparametric selection, and time-efficient sorting. All while preserving the viability of live cells for critical downstream applications.
– High Purity & Specificity: FACS isolates specific cell populations with up to 95–99% purity, even from complex samples. Fluorescence Activated Cell Sorting provides exceptionally high sorting accuracy because it classifies cells based on sensitive, clearly defined, high-resolution fluorescence signals. This is particularly important for subsequent applications, such as RNA sequencing, functional cell testing, and clonal expansion.
– Multiparametric Selection: By analyzing multiple features simultaneously such as surface markers, intracellular proteins, size, and granularity FACS can define and collect highly specific cell subsets with complex phenotypes in real time.
– High Viability: The process is gentle and non-destructive, preserving cell health and functional integrity. Sorted cells remain alive, making them suitable for sensitive applications like cell culture, proliferation studies, or therapeutic infusion.
– Rapid Throughput: Modern FACS instruments can analyze and sort several thousand to over 50,000 cells per second. This high throughput rate enables the processing of large sample volumes in a short amount of time, a significant advantage for clinical studies and high-throughput screening. Thanks to high-speed processing and sorting, even rare cellular events within a cell population can be reliably captured.
By merging speed, precision, and biocompatibility, FACS enables researchers to move from complex biological samples to ready-to-study, therapeutically relevant cell populations making it a cornerstone of modern molecular and translational science.
Click chemistry and FACS cell sorting: enhanced detection possibilities
Click chemistry labeling offers a highly specific and efficient way to tag biomolecules without disturbing cellular function. By using bioorthogonal reactions between azide and alkyne groups, it enables fast, low-background detection under mild conditions. This technique supports high-resolution imaging, multiplexing, and dynamic tracking of cellular processes like DNA synthesis through EdU based, protein turnover, and metabolism, making it ideal for both fixed and live-cell analysis.
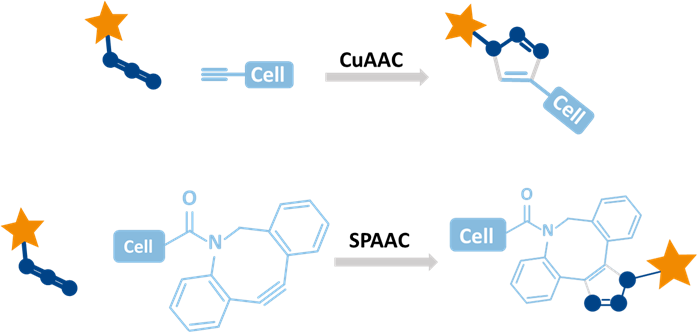
baseclick’s click chemistry reagents, like EdU and EU, integrate efficiently into FACS workflows by labeling DNA or RNA during synthesis without disrupting cell function. These modified nucleosides mimic natural bases, allowing incorporation by polymerases with no toxicity. Detection uses fast, specific click reactions under gentle conditions, preserving cell structure and viability. Whether in live or fixed cells, labeled samples yield clear fluorescence signals for analyzing proliferation, transcription, or cell cycle status all without compromising downstream FACS performance.
Comparison FACS cell sorting to magnetic cell sorting and other techniques
Cell isolation is a critical step in many biological and clinical workflows, and several methods exist depending on the required precision and downstream application. Techniques like FACS (Fluorescence Activated Cell Sorting), MACS (Magnetic Activated Cell Sorting), manual sorting (e.g., pipetting, adherence), and bulk enrichment (e.g., density gradient), each offer distinct advantages and limitations based on speed, specificity, and resolution. FACS offers the highest precision, sorting cells by multiple markers with single-cell resolution but it’s complex and slower. MACS is quicker and easier, ideal for bulk selection using one marker, though less detailed. Manual methods provide rough, low-precision separation. Bulk enrichment (like centrifugation) works well for initial sample prep but lacks fine phenotyping capability.
|
Method |
Principle | Precision | Throughput | Live cell Recovery | Multiparameter Capability |
Typical Use |
| FACS | Uses lasers to detect fluorescent markers and sort individual cells. | high | moderate | high | many markers simultaneously | Detailed phenotyping,
rare cell isolation |
| MACS | Uses magnetic beads bound to antibodies to isolate labeled cells | moderate | high | high | usually 1-2 markers | Fast positive/negative selection of cells |
| Manual Methods | Based on physical properties (e.g., size, adherence) | low | low | variable | none | Crude separation (e.g., adherent vs suspension) |
| Bulk enrichment | Separate based on physical properties like density (e.g., ficoll) | low | moderate | high | none | Enrich PBMCs or mononuclear cells broadly |
FACS (Fluorescence-Activated Cell Sorting) is the preferred technique when high specificity, detailed quantification, or rare cell isolation are essential. By using multiple fluorescent markers, FACS enables precise single-cell phenotyping, ideal for identifying immune subsets, stem cells, or cancer populations with overlapping markers. Its fluorescence-based readout allows for accurate quantification of expression levels and dynamic cell states, including proliferation using markers like EdU. Crucially, FACS excels in isolating rare cells, such as circulating tumor cells or antigen-specific lymphocytes, even at very low frequencies.
While FACS offers unmatched precision and depth, it is slower and more complex than alternatives. For routine or bulk selection (e.g., isolating CD4⁺ T cells), MACS is faster and more cost-effective, though less granular. Manual techniques and bulk enrichment are suitable for rough separation or sample preprocessing, but lack the resolution required for fine phenotyping.
In summary, FACS is the gold standard for detailed cellular analysis, especially in fields like immunology, oncology, stem cell research, and transcriptomics.
Future directions in FACS cell sorting technology
Automation and miniaturization
Recent innovations in high-throughput and microfluidic FACS technologies are revolutionizing cell sorting by enhancing speed, scalability, and data integration, while minimizing cellular stress. Modern high-throughput platforms like BD FACSymphony™ and Cytek Aurora apply multiple lasers and spectral unmixing to detect over 30 parameters per cell at rapid rates. Coupled with automated sampling, barcoding, and index sorting, they enable large-scale, multiparametric analysis and seamless transition to downstream workflows like single-cell genomics.
Microfluidic FACS systems offer a compact alternative, using microchannels and droplet-based sorting for gentle manipulation of rare or fragile cells. Their minimal sample requirements and integrated capabilities such as on-chip sequencing, lysis, or imaging, make them ideal for real-time functional assays and point-of-care diagnostics. Devices like On-chip Sort and Berkeley Lights’ Beacon® exemplify this shift toward portable, high-resolution platforms.
Together, these advancements blend precision, throughput, and versatility, supporting applications from immunophenotyping and CRISPR screens to rare cell analysis and live-cell functional profiling.
Integration with single-cell omics
FACS Cell Sorting is a powerful tool for preparing high-quality, cell-specific input for transcriptomics, epigenetics, and proteomics. Its ability to perform multiparameter, marker-based sorting at single-cell resolution ensures high purity and viability critical for sensitive downstream assays. For transcriptomics, FACS isolates distinct cell types or states with minimal contamination, enabling both single-cell RNA sequencing and precise bulk RNA profiling of rare populations. In epigenetic workflows, it maintains chromatin integrity while enriching for relevant cell types or nuclei, improving the accuracy of assays like ATAC-seq, ChIP-seq, and DNA methylation analysis. For proteomics, FACS delivers clean samples ideal for mass spectrometry or CyTOF, allowing detailed insights into protein expression and modifications, even in rare or complex tissues.
Its precision makes FACS the gold standard for multi-omics preparation, especially when dealing with heterogeneous samples, low-input requirements, or rare cells and it can be combined with techniques like index sorting to link phenotypic and molecular data in a single workflow.
Explore baseclick’s compatible tools for FACS cell sorting
baseclick GmbH offers a specialized suite of click chemistry-based labeling reagents and kits optimized for FACS workflows, particularly for detecting DNA synthesis, RNA transcription, and overall cell proliferation. These products are built around EdU and EU incorporation and feature bright, stable fluorophores that ensure strong signal clarity and multiplexing capability in flow cytometry. Their alkyne-modified nucleosides react with azide-linked dyes, available in various wavelengths like 488, 555, and 647 nm via highly specific click reactions, including copper-free SPAAC variants designed for sensitive or live-cell applications:
Beyond complete kits, baseclick also provides standalone fluorescent azides for custom FACS panels, compatible with EdU, EU, or other alkynyl-tagged biomolecules. Their formulations minimize cytotoxicity, preserve cell viability, and maintain fluorescence quality, making them especially suitable for high-resolution, multiparameter cell profiling in research settings like immunology, oncology, and stem cell biology:
Click-Compatible Fluorescent Dyes such as:
- Azide-Alexa Fluor 488, 555, 647
- Sulfo-Cy3, Cy5, TAMRA azides
Compatible with EdU/EU, or other alkyne-labeled biomolecules with high photostability and quantum yield and ideal for flow cytometry.

